Antimicrobial Resistance and Virulence Factors of Proteus mirabilis Isolated from Dog with Chronic Otitis Externa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Identification of Proteus mirabilis Isolates
2.2. Antimicrobial Susceptibility Test
2.3. Phenotypic Detection of Extended Spectrum Beta-Lactamase (ESBL)
2.4. Genotypic Detection of Antibiotic Resistance, Virulence, and Highly Conserved Genes
2.5. Phylogenic Investigation of P. mirabilis Isolates
3. Results
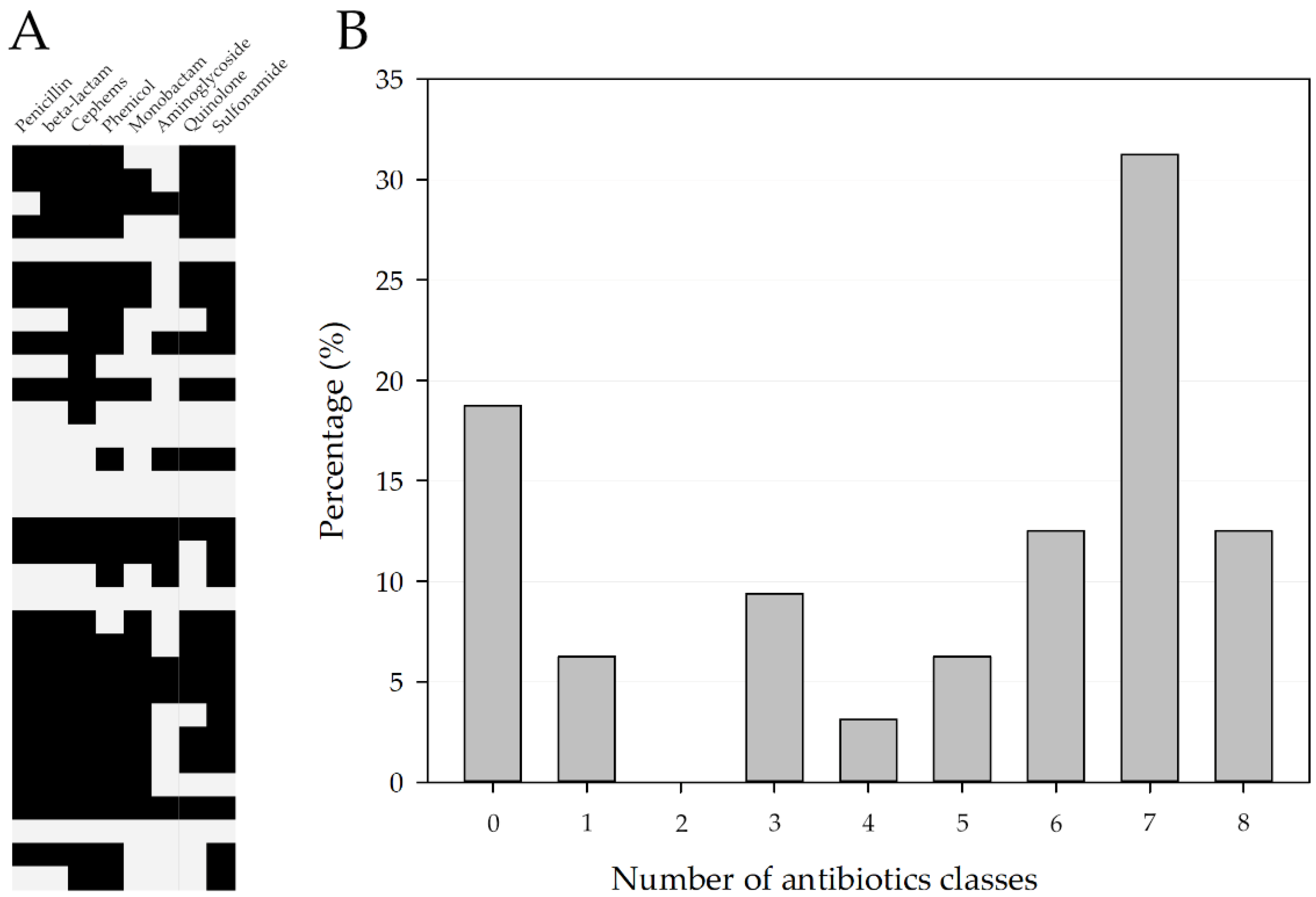
3.1. Antimicrobial Susceptibility Profile
3.2. Genotypic Description of Clinical Isolates of P. mirabilis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- August, J.R. Otitis externa: A disease of multifactorial etiology. Vet. Clin. N. Am. Small Anim. 1988, 18, 731–742. [Google Scholar] [CrossRef]
- McKeever, P.J. Otitis externa. Compend. Contin. Educ. Vet. 1996, 18, 759–773. [Google Scholar]
- Koutinas, A.F.; Saridomichelakis, M.N. Otitis externa in the dog and the eat. J. Hell. Vet. Med. 1998, 49, 251–252. [Google Scholar] [CrossRef] [Green Version]
- Angus, J.C. Otic cytology in health and disease. Vet. Clin. N. Am. Small Anim. Pract. 2004, 34, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Cole, L.K. Otoscopic evaluation of the ear canal. Vet. Clin. N. Am. Small Anim. Pract. 2004, 34, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Miller, W.H.; Griffin, C.E. External ear diseases. In Muller and Kirk’s Small Animal Dermatology, 6th ed.; Kersey, R., DiBerardino, C., Eds.; WB Saunders: Philadelphia, PA, USA, 2001; pp. 1203–1235. [Google Scholar]
- Rosser, E.J. Causes of otitis externa. Vet. Clin. Small Anim. Pract. 2004, 34, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Saridomichelakis, M.N. Strategies in the treatment of chronic or recurrent canine otitis externa-media. In Proceedings of the 20th Annual Congress of the European Society of Veterinary Dermatology-European College of Veterinary Dermatology, Chalkdiki, Greece, 8–10 September 2005; pp. 99–105. [Google Scholar]
- Saridomichelakis, M.N.; Farmaki, R.; Leontides, L.S.; Koutinas, A.F. Aetiology of canine otitis externa: A retrospective study of 100 cases. Vet. Dermatol. 2007, 18, 341–347. [Google Scholar] [CrossRef]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Goossens, H.; Ferech, M.; Vander Stichele, R.; Elseviers, M.; ESAC Project Group. Outpatient antibiotic use in Europe and association with resistance: A cross-national database study. Lancet 2005, 365, 579–587. [Google Scholar] [CrossRef]
- Dewulf, J.; Catry, B.; Timmerman, T.; Opsomer, G.; de Kruif, A.; Maes, D. Tetracycline-resistance in lactose-positive enteric coliforms originating from Belgian fattening pigs: Degree of resistance, multiple resistance and risk factors. Prev. Vet. Med. 2007, 78, 339–351. [Google Scholar] [CrossRef]
- Magalhães, R.J.S.; Loeffler, A.; Lindsay, J.; Rich, M.; Roberts, L.; Smith, H.; Lloyd, D.H.; Pfeiffer, D.U. Risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infection in dogs and cats: A case-control study. Vet. Res. 2010, 41, 55. [Google Scholar] [CrossRef]
- Burow, E.; Simoneit, C.; Tenhagen, B.A.; Käsbohrer, A. Oral antimicrobials increase antimicrobial resistance in porcine E. coli–A systematic review. Prev. Vet. Med. 2014, 113, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef] [Green Version]
- Van Duijkeren, E.; Schink, A.K.; Roberts, M.C.; Wang, Y.; Schwarz, S. Mechanisms of bacterial resistance to antimicrobial agents. Microbiol. Spectr. 2018, 6, 6-2. [Google Scholar] [CrossRef]
- Umber, J.K.; Bender, J.B. Pets and antimicrobial resistance. Vet. Clin. N. Am. Small Anim. Pract. 2009, 39, 279–292. [Google Scholar] [CrossRef]
- Zhang, X.F.; Doi, Y.; Huang, X.; Li, H.Y.; Zhong, L.L.; Zeng, K.J.; Zhang, Y.-F.; Patil, S.; Tian, G.B. Possible transmission of mcr-1–harboring Escherichia coli between companion animals and human. Emerg. Infect. Dis. 2016, 22, 1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, B.J.; Hungate, B.A.; Price, L.B. Food-animal production and the spread of antibiotic resistance: The role of ecology. Front. Ecol. Environ. 2017, 15, 309–318. [Google Scholar] [CrossRef]
- Odensvik, K.; Grave, K.; Greko, C. Antibacterial drugs prescribed for dogs and cats in Sweden and Norway 1990–1998. Acta Vet. Scand. 2001, 42, 1–10. [Google Scholar] [CrossRef]
- Middlemiss, C. Encouraging responsible antibiotic use by pet owners. Vet. Rec. 2018, 182, 410. [Google Scholar] [CrossRef] [PubMed]
- Somayaji, R.; Priyantha, M.A.R.; Rubin, J.E.; Church, D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: Report of 24 cases. Diagn. Microbiol. Infect. Dis. 2016, 85, 471–476. [Google Scholar] [CrossRef]
- Lozano, C.; Rezusta, A.; Ferrer, I.; Pérez-Laguna, V.; Zarazaga, M.; Ruiz-Ripa, L.; Revillo, M.J.; Torres, C. Staphylococcus pseudintermedius human infection cases in Spain: Dog-to-human transmission. Vector Borne Zoonotic Dis. 2017, 17, 268–270. [Google Scholar] [CrossRef]
- Robb, A.R.; Wright, E.D.; Foster, A.M.; Walker, R.; Malone, C. Skin infection caused by a novel strain of Staphylococcus pseudintermedius in a Siberian husky dog owner. JMM Case Rep. 2017, 4, jmmcr005087. [Google Scholar] [CrossRef] [PubMed]
- Battersby, I. Using antibiotics responsibly in companion animals. InPractice 2014, 36, 106–118. [Google Scholar] [CrossRef]
- PFMA Survey Confirms Stable Pet-Owning Population. Available online: https://www.vettimes.co.uk/news/pfma-survey-confirms-stable-pet-owning-population/ (accessed on 12 June 2020).
- Armbruster, C.E.; Mobley, H.L.; Pearson, M.M. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Wenner, J.J.; Rettger, L.F. A systematic study of the Proteus group of bacteria. J. Bacteriol. 1919, 4, 331–353. [Google Scholar] [CrossRef] [Green Version]
- Armbruster, C.E.; Mobley, H.L. Merging mythology and morphology: The multifaceted lifestyle of Proteus mirabilis. Nat. Rev. Microbiol. 2012, 10, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.W.; Tenney, J.H.; Hoopes, J.M.; Muncie, H.L.; Anthony, W.C. A prospective microbiologic study of bacteriuria in patients with chronic indwelling urethral catheters. J. Infect. Dis. 1982, 146, 719–723. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L.; Warren, J.W. Urease-positive bacteriuria and obstruction of long-term urinary catheters. J. Clin. Microbiol. 1987, 25, 2216–2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breitenbucher, R.B. Bacterial changes in the urine samples of patients with long-term indwelling catheters. Arch. Intern. Med. 1984, 144, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.Á.; Stickler, D.J.; Mobley, H.L.T.; Shirtliff, M.E. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 2008, 21, 26–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolle, L.E. Catheter-related urinary tract infection. Drugs Aging 2005, 22, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, C.E.; Prenovost, K.; Mobley, H.L.; Mody, L. How often do clinically diagnosed catheter-associated urinary tract infections in nursing homes meet standardized criteria? J. Am. Geriatr. Soc. 2017, 65, 395–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffith, D.P.; Musher, D.Á. Urease: Principal cause of infection stones. In Urolithiasis Research; Fleisch, H., Robertson, W.G., Smith, L.H., Vahlensieck, W., Eds.; Springer: Boston, MA, USA, 1976; pp. 451–454. [Google Scholar]
- Li, X.; Zhao, H.; Lockatell, C.V.; Drachenberg, C.B.; Johnson, D.E.; Mobley, H.L. Visualization of Proteus mirabilis within the matrix of urease-induced bladder stones during experimental urinary tract infection. Infect. Immun. 2002, 70, 389–394. [Google Scholar] [CrossRef] [Green Version]
- Foxman, B.; Brown, P. Epidemiology of urinary tract infections: Transmission and risk factors, incidence, and costs. Infect. Dis. Clin. N. Am. 2003, 17, 227–241. [Google Scholar] [CrossRef]
- Kim, B.N.; Kim, N.J.; Kim, M.N.; Kim, Y.S.; Woo, J.H.; Ryu, J. Bacteraemia due to tribe Proteeae: A review of 132 cases during a decade (1991–2000). Scand. J. Infect. Dis. 2003, 35, 98–103. [Google Scholar] [PubMed]
- Watanakunakorn, C.; Perni, S.C. Proteus mirabilis bacteremia: A review of 176 cases during 1980–1992. Scand. J. Infect. Dis. 1994, 26, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Daniels, K.R.; Lee, G.C.; Frei, C.R. Trends in catheter-associated urinary tract infections among a national cohort of hospitalized adults, 2001–2010. Am. J. Infect. Control. 2014, 42, 17–22. [Google Scholar] [PubMed]
- Hooton, T.M.; Bradley, S.F.; Cardenas, D.D.; Colgan, R.; Geerlings, S.E.; Rice, J.C.; Saint, S.; Schaeffer, A.J.; Tambayh, P.A.; Tenke, P.; et al. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin. Infect. Dis. 2010, 50, 625–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hijazin, M.; Alber, J.; Lämmler, C.; Weitzel, T.; Hassan, A.A.; Timke, M.; Kostrzewa, M.; Prenger-Berninghoff, E.; Zschöck, M. Identification of Trueperella (Arcanobacterium) bernardiae by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis and by species-specific PCR. J. Med. Microbiol. 2012, 61, 457–459. [Google Scholar]
- Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; M100; CLSI: Wayne, PA, USA, 2020; ISBN 1562387855. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Peng, C.; Zhang, G.; Shen, Y.; Zhang, Y.; Liu, C.; Liu, M.; Wang, F. Prevalence and characteristics of multidrug-resistant Proteus mirabilis from broiler farms in Shandong Province, China. Poult. Sci. 2022, 101, 101710. [Google Scholar] [CrossRef] [PubMed]
- Algammal, A.M.; Hashem, H.R.; Alfifi, K.J.; Hetta, H.F.; Sheraba, N.S.; Ramadan, H.; El-Tarabili, R.M. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 2021, 11, 9476. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- De Martino, L.; Nocera, F.P.; Mallardo, K.; Nizza, S.; Masturzo, E.; Fiorito, F.; Lovane, G.; Catalanotti, P. An update on microbiological causes of canine otitis externa in Campania Region, Italy. Asian Pac. J. Trop. Biomed. 2016, 6, 384–389. [Google Scholar] [CrossRef] [Green Version]
- Miller, W.H.; Griffin, C.E.; Campbell, K.L. Diseases of eyelids, claws, anal sacs, and ears. In Muller & Kirk’s Small Animal Dermatology, 7th ed.; Elsevier Inc.: St. Louis, MO, USA, 2013; pp. 724–773. [Google Scholar]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Microbiological effects of sublethal levels of antibiotics. Nat. Rev. Microbiol. 2014, 12, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Choi, S.M.; Park, S.H.; Lee, D.G.; Choi, J.H.; Yoo, J.H. Amikacin therapy for urinary tract infections caused by extended-spectrum β-lactamase-producing Escherichia coli. Korean J. Intern. Med. 2016, 31, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yee, R.; Dien Bard, J.; Simner, P.J. The genotype-to-phenotype dilemma: How should laboratories approach discordant susceptibility results? J. Clin. Microbiol. 2021, 59, e00138-20. [Google Scholar] [CrossRef]
- Swihart, K.G.; Welch, R.A. Cytotoxic activity of the Proteus hemolysin HpmA. Infect. Immun. 1990, 58, 1861–1869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Nielubowicz, G.; Mobley, H. Hostpathogen interactions in urinary tract infections. Nat. Rev. Urol. 2010, 7, 430–441. [Google Scholar] [CrossRef]
- Thomas, J.; Linton, S.; Corum, L.; Slone, W.; Okel, T.; Percival, S.L. The affect of pH and bacterial phenotypic state on antibiotic efficacy. Int. Wound J. 2021, 9, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Belas, R.; Schneider, R.; Melch, M. Characterization of Proteus mirabilis precocious swarming mutants: Identification of rsbA, encoding a regulator of swarming behavior. J. Bacteriol. Res. 1998, 180, 6126–6139. [Google Scholar] [CrossRef]
- Walker, K.E.; Moghaddame-Jafari, S.; Lockatell, C.V.; Johnson, D.; Belas, R. ZapA, the IgA-degrading metalloprotease of Proteus mirabilis, is a virulence factor expressed specifically in swarmer cells. Mol. Microbiol. 1999, 32, 825–836. [Google Scholar] [CrossRef]
- Belas, R.; Manos, J.; Suvanasuthi, R. Proteus mirabilis ZapA metalloprotease degrades a broad spectrum of substrates, including antimicrobial peptides. Infect. Immun. 2004, 72, 5159–5167. [Google Scholar] [CrossRef] [Green Version]
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.M.; Bechinger, B.; Naas, T. Antimicrobial peptides: A potent alternative to antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pathophysiol. 2017, 8, 162. [Google Scholar] [CrossRef]

| Antibiotics | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| AMP | 12 (38%) | 1 (3%) | 19 (59%) |
| AMC | 12 (37%) | 20 (63%) | |
| CN | 25 (78%) | 7 (22%) | |
| ATM | 14 (44%) | 2 (6%) | 16 (50%) |
| KZ | 8 (25%) | 24 (75%) | |
| AK | 27 (84%) | 5 (16%) | |
| CIP | 8 (25%) | 7 (22%) | 17 (53%) |
| SXT | 9 (28%) | 23 (72%) | |
| C | 8 (25%) | 1 (3%) | 23 (72%) |
| CZA | 16 (50%) | 16 (50%) | |
| FEP | 12 (38%) | 2 (6%) | 18 (56%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, J.; Yang, M.-H.; Ko, H.-J.; Kim, S.-G.; Park, C.; Park, S.-C. Antimicrobial Resistance and Virulence Factors of Proteus mirabilis Isolated from Dog with Chronic Otitis Externa. Pathogens 2022, 11, 1215. https://doi.org/10.3390/pathogens11101215
Kwon J, Yang M-H, Ko H-J, Kim S-G, Park C, Park S-C. Antimicrobial Resistance and Virulence Factors of Proteus mirabilis Isolated from Dog with Chronic Otitis Externa. Pathogens. 2022; 11(10):1215. https://doi.org/10.3390/pathogens11101215
Chicago/Turabian StyleKwon, Jun, Myoung-Hwan Yang, Hyoung-Joon Ko, Sang-Guen Kim, Chul Park, and Se-Chang Park. 2022. "Antimicrobial Resistance and Virulence Factors of Proteus mirabilis Isolated from Dog with Chronic Otitis Externa" Pathogens 11, no. 10: 1215. https://doi.org/10.3390/pathogens11101215
APA StyleKwon, J., Yang, M. -H., Ko, H. -J., Kim, S. -G., Park, C., & Park, S. -C. (2022). Antimicrobial Resistance and Virulence Factors of Proteus mirabilis Isolated from Dog with Chronic Otitis Externa. Pathogens, 11(10), 1215. https://doi.org/10.3390/pathogens11101215






