High-Specificity Test Algorithm for Bovine Tuberculosis Diagnosis in African Buffalo (Syncerus caffer) Herds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Single Comparative Intradermal Tuberculin Tests
2.3. Cytokine Release Assays
2.4. Bovine TB Test Interpretation
2.5. Mycobacterial Culture and Speciation
2.6. Data Analyses
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnot, L.F.; Michel, A. Challenges for Controlling Bovine Tuberculosis in South Africa. Onderstepoort J. Vet. Res. 2020, 87, e1–e8. [Google Scholar] [CrossRef] [PubMed]
- Borham, M.; Oreiby, A.; El-Gedawy, A.; Hegazy, Y.; Khalifa, H.O.; Al-Gaabary, M.; Matsumoto, T. Review on Bovine Tuberculosis: An Emerging Disease Associated with Multidrug-Resistant Mycobacterium Species. Pathogens 2022, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Van Pittius, N.C.G.; Perrett, K.D.; Michel, A.L.; Keet, D.F.; Hlokwe, T.; Streicher, E.M.; Warren, R.M.; van Helden, P.D. Infection of African Buffalo (Syncerus caffer) by Oryx Bacillus, a Rare Member of the Antelope Clade of the Mycobacterium tuberculosis Complex. J. Wildl. Dis. 2012, 48, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Borham, M.; Oreiby, A.; El-Gedawy, A.; Hegazy, Y.; Hemedan, A.; Al-Gaabary, M. Abattoir Survey of Bovine Tuberculosis in Tanta, Centre of the Nile Delta, with in Silico Analysis of Gene Mutations and Protein–Protein Interactions of the Involved Mycobacteria. Transbound. Emerg. Dis. 2022, 69, 434–450. [Google Scholar] [CrossRef]
- De Vos, V.; Bengis, R.G.; Kriek, N.P.J.; Michel, A.; Keet, D.F.; Raath, J.P.; Huchzermeyer, H.F.K.A. The Epidemiology of Tuberculosis in Free-Ranging African Buffalo (Syncerus caffer) in the Kruger National Park, South Africa. Onderstepoort J. Vet. Res. 2001, 68, 119–130. [Google Scholar]
- Department of Agriculture, Forestry and Fisheries (DAFF). Bovine Tuberculosis Manual 2016. Available online: https://www.nda.agric.za/vetweb/pamphlets&Information/Policy/Tuberculosis%20in%20Cattle%20Interim%20Manual%20for%20the%20Veterinarian%20&%20AHT%20-%20Sept2....pdf (accessed on 7 October 2022).
- Department of Agriculture, Land Reform and Rural Development (DALRRD). Veterinary Procedudaral Notice for Buffalo Disease Risk Management in South Africa 2017. Available online: https://www.dalrrd.gov.za/vetweb/pamphlets&Information/Policy/Buffalo%20Disease%20Risk%20Management%20VPN_Signed%202017-02-17.pdf (accessed on 5 October 2022).
- de la Rua-Domenech, R.; Goodchild, A.T.; Vordermeier, H.M.; Hewinson, R.G.; Christiansen, K.H.; Clifton-Hadley, R.S. Ante Mortem Diagnosis of Tuberculosis in Cattle: A Review of the Tuberculin Tests, γ-Interferon Assay and Other Ancillary Diagnostic Techniques. Res. Vet. Sci. 2006, 81, 190–210. [Google Scholar] [CrossRef]
- Vordermeier, H.M.; Brown, J.; Cockle, P.J.; Franken, W.P.J.; Arend, S.M.; Ottenhoff, T.H.M.; Jahans, K.; Hewinson, R.G. Assessment of Cross-Reactivity between Mycobacterium bovis and M. kansasii ESAT-6 and CFP-10 at the T-Cell Epitope Level. Clin. Vaccine Immunol. 2007, 14, 1203–1209. [Google Scholar] [CrossRef] [Green Version]
- Bernitz, N.; Kerr, T.; Goosen, W.; Chileshe, J.; Higgit, R.; Roos, E.; Meiring, C.; Gumbo, R.; de Waal, C.; Clarke, C.; et al. Review of Diagnostic Tests for Detection of Mycobacterium bovis Infection in South African Wildlife. Front. Vet. Sci. 2021, 8, 588697. [Google Scholar] [CrossRef]
- Parsons, S.D.C.; Cooper, D.; McCall, A.J.; McCall, W.A.; Streicher, E.M.; le Maitre, N.C.; Müller, A.; Gey van Pittius, N.C.; Warren, R.M.; van Helden, P.D. Modification of the QuantiFERON-TB Gold (In-Tube) Assay for the Diagnosis of Mycobacterium bovis Infection in African Buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2011, 142, 113–118. [Google Scholar] [CrossRef]
- Smith, K.; Bernitz, N.; Goldswain, S.; Cooper, D.V.; Warren, R.M.; Goosen, W.J.; Miller, M.A. Optimized Interferon-Gamma Release Assays for Detection of Mycobacterium bovis Infection in African Buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2021, 231, 110163. [Google Scholar] [CrossRef]
- Bernitz, N.; Kerr, T.J.; Goosen, W.J.; Clarke, C.; Higgitt, R.; Roos, E.O.; Cooper, D.V.; Warren, R.M.; van Helden, P.D.; Parsons, S.D.C.; et al. Parallel Measurement of IFN-γ and IP-10 in QuantiFERON®-TB Gold (QFT) Plasma Improves the Detection of Mycobacterium bovis Infection in African Buffaloes (Syncerus caffer). Prev. Vet. Med. 2019, 169, 104700. [Google Scholar] [CrossRef] [PubMed]
- Lyashchenko, K.P.; Sridhara, A.A.; Johnathan-Lee, A.; Sikar-Gang, A.; Lambotte, P.; Esfandiari, J.; Bernitz, N.; Kerr, T.J.; Miller, M.A.; Waters, W.R. Differential Antigen Recognition by Serum Antibodies from Three Bovid Hosts of Mycobacterium bovis Infection. Comp. Immunol. Microbiol. Infect. Dis. 2020, 69, 101424. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.L.; Cooper, D.; Jooste, J.; de Klerk, L.M.; Jolles, A. Approaches Towards Optimising the Gamma Interferon Assay for Diagnosing Mycobacterium bovis Infection in African Buffalo (Syncerus caffer). Prev. Vet. Med. 2011, 98, 142–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.; Bernitz, N.; Cooper, D.; Kerr, T.J.; de Waal, C.R.; Clarke, C.; Goldswain, S.; McCall, W.; McCall, A.; Cooke, D.; et al. Optimisation of the Tuberculin Skin Test for Detection of Mycobacterium bovis in African Buffaloes (Syncerus caffer). Prev. Vet. Med. 2021, 188, 105254. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, A.O.; Gormley, E.; Gcebe, N.; Fosgate, G.T.; Conan, A.; Aagaard, C.; Michel, A.L.; Rutten, V.P.M.G. Cross Reactive Immune Responses in Cattle Arising from Exposure to Mycobacterium bovis and Non-Tuberculous Mycobacteria. Prev. Vet. Med. 2018, 152, 16–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varela-Castro, L.; Barral, M.; Arnal, M.C.; Fernández de Luco, D.; Gortázar, C.; Garrido, J.M.; Sevilla, I.A. Beyond Tuberculosis: Diversity and Implications of Non-tuberculous Mycobacteria at the Wildlife-Livestock Interface. Transbound. Emerg. Dis. 2022, 69, e2978–e2993. [Google Scholar] [CrossRef] [PubMed]
- Bernitz, N.; Kerr, T.J.; de Waal, C.; Cooper, D.V.; Warren, R.M.; Van Helden, P.D.; Parsons, S.D.C.; Miller, M.A. Test Characteristics of Assays to Detect Mycobacterium bovis Infection in High-Prevalence African Buffalo (Syncerus caffer) Herds. J. Wildl. Dis. 2020, 56, 462–465. [Google Scholar] [CrossRef]
- Goosen, W.J.; Cooper, D.; Miller, M.A.; Van Helden, P.D.; Parsons, S.D.C. IP-10 Is a Sensitive Biomarker of Antigen Recognition in Whole-Blood Stimulation Assays Used for the Diagnosis of Mycobacterium bovis Infection in African Buffaloes (Syncerus caffer). Clin. Vaccine Immunol. 2015, 22, 974–978. [Google Scholar] [CrossRef] [Green Version]
- Goosen, W.J.; Miller, M.A.; Chegou, N.N.; Cooper, D.; Warren, R.M.; van Helden, P.D.; Parsons, S.D.C. Agreement between Assays of Cell-Mediated Immunity Utilizing Mycobacterium bovis-Specific Antigens for the Diagnosis of Tuberculosis in African Buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2014, 160, 133–138. [Google Scholar] [CrossRef]
- Clarke, C.; Kerr, T.J.; Warren, R.M.; Kleynhans, L.; Miller, M.A.; Goosen, W.J. Identification and Characterisation of Nontuberculous Mycobacteria in African Buffaloes (Syncerus caffer), South Africa. Microorganisms 2022, 10, 1861. [Google Scholar] [CrossRef]
- Adékambi, T.; Colson, P.; Drancourt, M. RpoB-Based Identification of Nonpigmented and Late-Pigmenting Rapidly Growing Mycobacteria. J. Clin. Microbiol. 2003, 41, 5699–5708. [Google Scholar] [CrossRef] [PubMed]
- Telenti, A.; Marchesi, F.; Balz, M.; Bally, F.; Bottger, E.C.; Bodmer, T. Rapid Identification of Mycobacteria to the Species Level by Polymerase Chain Reaction and Restriction Enzyme Analysis. J. Clin. Microbiol. 1993, 31, 175–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- To, K.; Cao, R.; Yegiazaryan, A.; Owens, J.; Venketaraman, V. General Overview of Nontuberculous Mycobacteria Opportunistic Pathogens: Mycobacterium avium and Mycobacterium abscessus. J. Clin. Med. 2020, 9, 2541. [Google Scholar] [CrossRef] [PubMed]
- Warren, R.M.; Gey Van Pittius, N.C.; Barnard, M.; Hesseling, A.; Engelke, E.; De Kock, M.; Gutierrez, M.C.; Chege, G.K.; Victor, T.C.; Hoal, E.G.; et al. Differentiation of Mycobacterium tuberculosis Complex by PCR Amplification of Genomic Regions of Difference. Int. J. Tuberc. Lung Dis. 2006, 10, 818–822. [Google Scholar]
- Scherrer, S.; Landolt, P.; Friedel, U.; Stephan, R. Distribution and Expression of esat-6 and cfp-10 in Non-Tuberculous Mycobacteria Isolated from Lymph Nodes of Slaughtered Cattle in Switzerland. J. Vet. Diagn. Investig. 2019, 31, 217–221. [Google Scholar] [CrossRef] [Green Version]
- Bernitz, N.; Goosen, W.J.; Clarke, C.; Kerr, T.J.; Higgitt, R.; Roos, E.O.; Cooper, D.V.; Warren, R.M.; van Helden, P.D.; Parsons, S.D.C.; et al. Parallel Testing Increases Detection of Mycobacterium bovis-Infected African Buffaloes (Syncerus caffer). Vet. Immunol. Immunopathol. 2018, 204, 40–43. [Google Scholar] [CrossRef]
- Van Ingen, J.; De Zwaan, R.; Dekhuijzen, R.; Boeree, M.; Van Soolingen, D. Region of Difference 1 in Nontuberculous Mycobacterium Species Adds a Phylogenetic and Taxonomical Character. J. Bacteriol. 2009, 191, 5865–5867. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.Y.; Reddy, T.B.K.; Arend, S.M.; Friggen, A.H.; Franken, K.L.M.C.; van Meijgaarden, K.E.; Verduyn, M.J.C.; Schoolnik, G.K.; Klein, M.R.; Ottenhoff, T.H.M. Cross-Reactive Immunity to Mycobacterium tuberculosis DosR Regulon-Encoded Antigens in Individuals Infected with Environmental, Nontuberculous Mycobacteria. Infect. Immun. 2009, 77, 5071–5079. [Google Scholar] [CrossRef] [Green Version]
- Malama, S.; Munyeme, M.; Mwanza, S.; Muma, J.B. Isolation and Characterization of Nontuberculous Mycobacteria from Humans and Animals in Namwala District of Zambia. BMC Res. Notes 2014, 7, 622. [Google Scholar] [CrossRef] [Green Version]
- Hermansen, T.S.; Thomsen, V.Ø.; Lillebaek, T.; Ravn, P. Non-Tuberculous Mycobacteria and the Performance of Interferon Gamma Release Assays in Denmark. PLoS ONE 2014, 9, e93986. [Google Scholar] [CrossRef]
| Test | False Positive | True Negative | Specificity (95% Confidence Interval) |
|---|---|---|---|
| IGRA a (n = 115) | 11 (9.6%) | 104 (90.4%) | 90.4% (83.5–95.1%) |
| IPRA b (n = 115) | 22 (19.1%) | 93 (80.9%) | 80.9% (72.5–87.6%) |
| SCITT (n = 69) | 0 (0%) | 69 (100%) | 100% (94.8–100%) |
| Parallel testing (IGRA a, IPRA b) | 31 (27%) | 84 (73%) | 73.0% (64.0–80.9%) |
| Serial testing (IGRA a, IPRA b) | 2 (1.7%) | 113 (98.3%) | 98.3% (93.9–99.8%) |
| Serial testing (IGRA a, IPRA b, SCITT) | 0 (0%) | 69 (100%) | 100% (94.8–100%) |
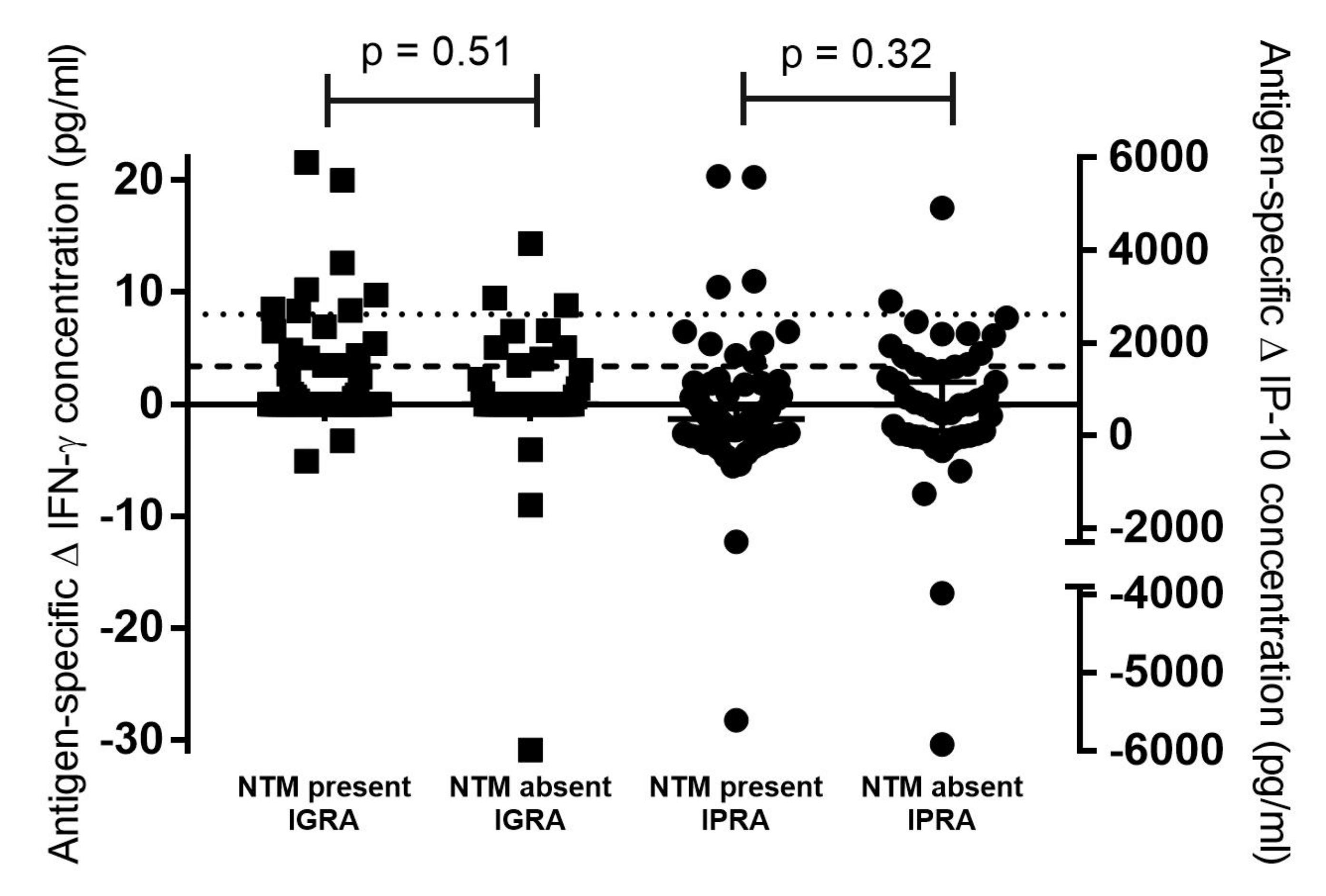
| NTM Present | NTM Absent | p-Value (Fisher’s Exact Test) | |
|---|---|---|---|
| IGRA a test-positive | 8/11 (73%) | 3/11 (27%) | 0.5 |
| IGRA a test-negative | 61/104 (59%) | 43/104 (41%) | |
| IPRA b test-positive | 10/22 (45%) | 12/22 (55%) | 0.15 |
| IPRA b test-negative | 59/93 (63%) | 34/93 (37%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, C.; Bernitz, N.; Goosen, W.J.; Miller, M.A. High-Specificity Test Algorithm for Bovine Tuberculosis Diagnosis in African Buffalo (Syncerus caffer) Herds. Pathogens 2022, 11, 1393. https://doi.org/10.3390/pathogens11121393
Clarke C, Bernitz N, Goosen WJ, Miller MA. High-Specificity Test Algorithm for Bovine Tuberculosis Diagnosis in African Buffalo (Syncerus caffer) Herds. Pathogens. 2022; 11(12):1393. https://doi.org/10.3390/pathogens11121393
Chicago/Turabian StyleClarke, Charlene, Netanya Bernitz, Wynand J. Goosen, and Michele A. Miller. 2022. "High-Specificity Test Algorithm for Bovine Tuberculosis Diagnosis in African Buffalo (Syncerus caffer) Herds" Pathogens 11, no. 12: 1393. https://doi.org/10.3390/pathogens11121393
APA StyleClarke, C., Bernitz, N., Goosen, W. J., & Miller, M. A. (2022). High-Specificity Test Algorithm for Bovine Tuberculosis Diagnosis in African Buffalo (Syncerus caffer) Herds. Pathogens, 11(12), 1393. https://doi.org/10.3390/pathogens11121393







