Application of Fluorescence In Situ Hybridization (FISH) in Oral Microbial Detection
Abstract
:1. Introduction
2. Research Progress of FISH
2.1. Development of FISH
2.2. Procedures and Principles of FISH and Its Variants Used for Oral Microbial Detection
2.2.1. FISH
2.2.2. CLASI-FISH
2.2.3. HiPR-FISH
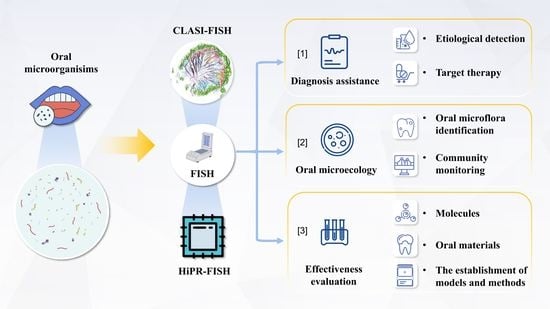
3. Research and Application of FISH in Oral Microbial Detection
3.1. Diagnosis Assistance
3.2. Oral Microecology
3.3. Effectiveness Evaluation
3.3.1. Evaluation of the Effectiveness of Molecules
3.3.2. Evaluation of the Bacteriostatic Effect of Oral Materials
3.3.3. Verification of the Establishment of Models and Methods
4. Discussion and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arweiler, N.B.; Netuschil, L. The Oral Microbiota. Adv. Exp. Med. Biol. 2016, 902, 45–60. [Google Scholar] [PubMed]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol. 2014, 60, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Wade, W.G. The oral microbiome in health and disease. Pharmacol. Res. 2013, 69, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Garg, P.K.; Dubey, A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018, 200, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Bor, B.; Agnello, M.; Shi, W.; He, X. Ecology of the Oral Microbiome: Beyond Bacteria. Trends Microbiol. 2017, 25, 362–374. [Google Scholar] [CrossRef] [Green Version]
- Xian, P.; Xuedong, Z.; Xin, X.; Yuqing, L.; Yan, L.; Jiyao, L.; Xiaoquan, S.; Shi, H.; Jian, X.; Ga, L. The Oral Microbiome Bank of China. Int. J. Oral Sci. 2018, 10, 6. [Google Scholar] [CrossRef]
- Welch, J.L.M.; Ramírez-Puebla, S.T.; Borisy, G.G. Oral Microbiome Geography: Micron-Scale Habitat and Niche. Cell Host Microbe 2020, 28, 160–168. [Google Scholar] [CrossRef]
- Scannapieco, F.A. Saliva-Bacterium Interactions in Oral Microbial Ecology. Crit. Rev. Oral Biol. Med. 1994, 5, 203–248. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Xu, T.; Huang, G.; Jiang, S.; Gu, Y.; Chen, F. Oral microbiomes: More and more importance in oral cavity and whole body. Protein Cell 2018, 9, 488–500. [Google Scholar] [CrossRef]
- Pascale, A.; Marchesi, N.; Marelli, C.; Coppola, A.; Luzi, L.; Govoni, S.; Giustina, A.; Gazzaruso, C. Microbiota and metabolic diseases. Endocrine 2018, 61, 357–371. [Google Scholar] [CrossRef]
- Mira, A. Oral Microbiome Studies: Potential Diagnostic and Therapeutic Implications. Adv. Dent. Res. 2018, 29, 71–77. [Google Scholar] [CrossRef]
- Prudent, E.; Raoult, D. Fluorescence in situ hybridization, a complementary molecular tool for the clinical diagnosis of infectious diseases by intracellular and fastidious bacteria. FEMS Microbiol. Rev. 2018, 43, 88–107. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Fröjd, V.; Linderbäck, P.; Wennerberg, A.; de Paz, L.C.; Svensäter, G.; Davies, J.R. Effect of nanoporous TiO2 coating and anodized Ca2+ modification of titanium surfaces on early microbial biofilm formation. BMC Oral Heal. 2011, 11, 8. [Google Scholar] [CrossRef] [Green Version]
- Guimaraes, N.M.; Azevedo, N.F.; Almeida, C. FISH Variants. Methods Mol. Biol. 2021, 2246, 17–33. [Google Scholar]
- Levsky, J.M.; Singer, R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef] [Green Version]
- Veselinyová, D.; Mašlanková, J.; Kalinová, K.; Mičková, H.; Mareková, M.; Rabajdová, M. Selected In Situ Hybridization Methods: Principles and Application. Molecules 2021, 26, 3874. [Google Scholar] [CrossRef]
- Gall, J.G.; Pardue, M.L. ormation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. USA 1969, 63, 378–383. [Google Scholar] [CrossRef] [Green Version]
- John, H.A.; Birnstiel, M.L.; Jones, K.W. RNA-DNA Hybrids at the Cytological Level. Nature 1969, 223, 582–587. [Google Scholar] [CrossRef]
- Beutner, E.H.; Holborow, E.J.; Johnson, G.D. A New Fluorescent Antibody Method: Mixed Antiglobulin Immunofluores-cence or Labelled Antigen Indirect Immunofluorescence Staining. Nature 1965, 208, 353–355. [Google Scholar] [CrossRef] [PubMed]
- Bauman, J.G.; Wiegant, J.; Borst, P.; Van Duijn, P. A new method for fluorescence microscopical localization of specific DNA sequences by in situ hybridi-zation of fluorochromelabelled RNA. Exp. Cell Res. 1980, 128, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Rudkin, G.T.; Stollar, B.D. High resolution detection of DNA–RNA hybrids in situ by indirect immunofluorescence. Nature 1977, 265, 472–473. [Google Scholar] [CrossRef] [PubMed]
- Zwirglmaier, K. Fluorescence in situ hybridisation (FISH)—The next generation. FEMS Microbiol. Lett. 2005, 246, 151–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, M.; Yang, B.; Cheng, Y.; Radda, J.S.D.; Chen, Y.; Liu, M.; Wang, S. ProbeDealer is a convenient tool for designing probes for highly multiplexed fluorescence in situ hybridization. Sci. Rep. 2020, 10, 22031. [Google Scholar] [CrossRef]
- Teixeira, H.; Sousa, A.L.; Azevedo, A.S. Bioinformatic Tools and Guidelines for the Design of Fluorescence In Situ Hy-bridization Probes. Methods Mol. Biol. 2021, 2246, 35–50. [Google Scholar]
- Yilmaz, L.S.; Parnerkar, S.; Noguera, D.R. mathFISH, a Web Tool That Uses Thermodynamics-Based Mathematical Models for In Silico Evaluation of Oligonucleotide Probes for Fluorescence In Situ Hybridization. Appl. Environ. Microbiol. 2011, 77, 1118–1122. [Google Scholar] [CrossRef] [Green Version]
- Hershberg, E.A.; Camplisson, C.K.; Close, J.L.; Attar, S.; Chern, R.; Liu, Y.; Akilesh, S.; Nicovich, P.R.; Beliveau, B.J. PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat. Methods 2021, 18, 937–944. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, T. Single Copy Oligonucleotide Fluorescence In Situ Hybridization Probe Design Platforms: Development, Application and Evaluation. Int. J. Mol. Sci. 2021, 22, 7124. [Google Scholar] [CrossRef]
- Zwirglmaier, K.; Ludwig, W.; Schleifer, K.H. Recognition of individual genes in a single bacterial cell by fluorescence in situ hybridization—RING-FISH. Mol. Microbiol. 2003, 51, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Dugan, L.C.; Pattee, M.S.; Williams, J.; Sorensen, K.; Bedford, J.S.; Christian, A.T. Polymerase chain reaction-based suppression of repetitive sequences in whole chromosome painting probes for FISH. Chromosom. Res. 2005, 13, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Pratscher, J.; Stichternoth, C.; Fichtl, K.; Schleifer, K.-H.; Braker, G. Application of Recognition of Individual Genes-Fluorescence In Situ Hybridization (RING-FISH) To Detect Nitrite Reductase Genes ( nirK ) of Denitrifiers in Pure Cultures and Environmental Samples. Appl. Environ. Microbiol. 2009, 75, 802–810. [Google Scholar] [CrossRef]
- Kawakami, S.; Hasegawa, T.; Imachi, H.; Yamaguchi, T.; Harada, H.; Ohashi, A.; Kubota, K. Detection of single-copy functional genes in prokaryotic cells by two-pass TSA-FISH with polynucleotide probes. J. Microbiol. Methods 2012, 88, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Moraru, C.; Lam, P.; Fuchs, B.M.; Kuypers, M.M.M.; Amann, R. GeneFISH—An in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ. Microbiol. 2010, 12, 3057–3073. [Google Scholar] [CrossRef]
- Barrero-Canosa, J.; Moraru, C.; Zeugner, L.; Fuchs, B.M.; Amann, R. Direct-geneFISH: A simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ. Microbiol. 2017, 19, 70–82. [Google Scholar] [CrossRef]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef]
- Fontenete, S.; Carvalho, D.; Guimarães, N.; Madureira, P.; Figueiredo, C.; Wengel, J.; Azevedo, N.F. Application of locked nucleic acid-based probes in fluorescence in situ hybridization. Appl. Microbiol. Biotechnol. 2016, 100, 5897–5906. [Google Scholar] [CrossRef]
- Geier, B.; Sogin, E.; Michellod, D.; Janda, M.; Kompauer, M.; Spengler, B.; Dubilier, N.; Liebeke, M. Spatial metabolomics of in situ host–microbe interactions at the micrometre scale. Nat. Microbiol. 2020, 5, 498–510. [Google Scholar] [CrossRef]
- Valm, A.M.; Welch, J.L.M.; Borisy, G.G. CLASI-FISH: Principles of combinatorial labeling and spectral imaging. Syst. Appl. Microbiol. 2012, 35, 496–502. [Google Scholar] [CrossRef] [Green Version]
- Behnam, F.; Vilcinskas, A.; Wagner, M.; Stoecker, K. A Straightforward DOPE (Double Labeling of Oligonucleotide Probes)-FISH (Fluorescence In Situ Hybridization) Method for Simultaneous Multicolor Detection of Six Microbial Populations. Appl. Environ. Microbiol. 2012, 78, 5138–5142. [Google Scholar] [CrossRef] [Green Version]
- Stoecker, K.; Dorninger, C.; Daims, H.; Wagner, M. Double Labeling of Oligonucleotide Probes for Fluorescence In Situ Hybridization (DOPE-FISH) Improves Signal Intensity and Increases rRNA Accessibility. Appl. Environ. Microbiol. 2010, 76, 922–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onozato, M.L.; Yapp, C.; Richardson, D.; Sundaresan, T.; Chahal, V.; Lee, J.; Sullivan, J.P.; Madden, M.W.; Shim, H.S.; Liebers, M.; et al. Highly Multiplexed Fluorescence in Situ Hybridization for in Situ Genomics. J. Mol. Diagn. 2019, 21, 390–407. [Google Scholar] [CrossRef] [PubMed]
- Petukhov, V.; Xu, R.J.; Soldatov, R.A.; Cadinu, P.; Khodosevich, K.; Moffitt, J.R.; Kharchenko, P.V. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 2021, 40, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Xia, C.; Close, J.L.; Zhang, M.; He, J.; Huang, Z.; Halpern, A.R.; Long, B.; Miller, J.A.; Lein, E.S. Conservation and divergence of cortical cell organization in human and mouse revealed by MERFISH. Science 2022, 377, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Lohoff, T.; Ghazanfar, S.; Missarova, A.; Koulena, N.; Pierson, N.; Griffiths, J.A.; Bardot, E.S.; Eng, C.-H.L.; Tyser, R.C.V.; Argelaguet, R.; et al. Integration of spatial and single-cell transcriptomic data elucidates mouse organogenesis. Nat. Biotechnol. 2021, 40, 74–85. [Google Scholar] [CrossRef]
- Wang, Y.; Eddison, M.; Fleishman, G.; Weigert, M.; Xu, S.; Wang, T.; Rokicki, K.; Goina, C.; Henry, F.E.; Lemire, A.L.; et al. EASI-FISH for thick tissue defines lateral hypothalamus spatio-molecular organization. Cell 2021, 184, 6361–6377.e24. [Google Scholar] [CrossRef]
- Egloff, S.; Melnychuk, N.; Da Silva, E.C.; Reisch, A.; Martin, S.; Klymchenko, A.S. Amplified Fluorescence in Situ Hybridization by Small and Bright Dye-Loaded Polymeric Nanoparticles. ACS Nano 2021, 16, 1381–1394. [Google Scholar] [CrossRef]
- Cui, C.; Shu, W.; Li, P. Fluorescence In situ Hybridization: Cell-Based Genetic Diagnostic and Research Applications. Front. Cell Dev. Biol. 2016, 4, 89. [Google Scholar] [CrossRef] [Green Version]
- Upendram, P.; Sahni, S.; Mohiuddin, K.; Poornima, S.; Gourishankar, B.; Vattam, K.K.; Boddala, P.; Jayashankar, E.; Mohiuddin, S.; Kamineni, V.; et al. Amplification of specific chromosomal regions assessed by fluorescent in situ hybridization on Pap smears to be added as screening tool for identifying women at risk of progressing to cervical cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Pauciullo, A.; Versace, C.; Perucatti, A.; Gaspa, G.; Li, L.-Y.; Yang, C.-Y.; Zheng, H.-Y.; Liu, Q.; Shang, J.-H. Oocyte aneuploidy rates in river and swamp buffalo types (Bubalus bubalis) determined by Multi-color Fluorescence In Situ Hybridization (M-FISH). Sci. Rep. 2022, 12, 8440. [Google Scholar] [CrossRef]
- Waminal, N.E.; Yang, T.-J.; In, J.-G.; Kim, H.H. Five-color fluorescence in situ hybridization system for karyotyping of Panax ginseng. Hortic. Environ. Biotechnol. 2020, 61, 869–877. [Google Scholar] [CrossRef]
- Ju, D.; Li, X.; Shi, Y.; Ma, Y.; Guo, L.; Wang, Y.; Ma, R.; Zhong, Y.; Zhang, Y.; Xue, F. Evaluation of the practical applications of fluorescence in situ hybridization in the prenatal diagnosis of positive noninvasive prenatal screenings. J. Matern. Neonatal Med. 2021, 35, 7422–7429. [Google Scholar] [CrossRef]
- Ha, J.; Cho, H.; Lee, T.G.; Shin, S.; Chung, H.; Jang, J.E.; Kim, S.-J.; Cheong, J.-W.; Lee, S.-T.; Kim, J.S.; et al. Cytogenetic testing by fluorescence in situ hybridization is improved by plasma cell sorting in multiple myeloma. Sci. Rep. 2022, 12, 8287. [Google Scholar] [CrossRef]
- Pereira, A.C.; Tenreiro, A.; Cunha, M.V. When FLOW-FISH met FACS: Combining multiparametric, dynamic approaches for microbial single-cell research in the total environment. Sci. Total Environ. 2021, 806, 150682. [Google Scholar] [CrossRef]
- Querido, E.; Dekakra-Bellili, L.; Chartrand, P. RNA fluorescence in situ hybridization for high-content screening. Methods 2017, 126, 149–155. [Google Scholar] [CrossRef]
- Brown, J.M.; De Ornellas, S.; Parisi, E.; Schermelleh, L.; Buckle, V.J. RASER-FISH: Non-denaturing fluorescence in situ hybridization for preservation of three-dimensional interphase chromatin structure. Nat. Protoc. 2022, 17, 1306–1331. [Google Scholar] [CrossRef]
- Al-Ahmad, A.; Roth, D.; Wolkewitz, M.; Wiedmann-Al-Ahmad, M.; Follo, M.; Ratka-Krüger, P.; Deimling, D.; Hellwig, E.; Hannig, C. Change in diet and oral hygiene over an 8-week period: Effects on oral health and oral biofilm. Clin. Oral Investig. 2009, 14, 391–396. [Google Scholar] [CrossRef]
- Alraies, A.; Canetta, E.; Waddington, R.J.; Moseley, R.; Sloan, A.J. Discrimination of Dental Pulp Stem Cell Regenerative Heterogeneity by Single-Cell Raman Spectroscopy. Tissue Eng. Part C Methods 2019, 25, 489–499. [Google Scholar] [CrossRef]
- Brinig, M.M.; Lepp, P.W.; Ouverney, C.; Armitage, G.C.; Relman, D.A. Prevalence of Bacteria of Division TM7 in Human Subgingival Plaque and Their Association with Disease. Appl. Environ. Microbiol. 2003, 69, 1687–1694. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wong, W.K.; Seneviratne, J.C.; Huang, S.; McGrath, C.; Hagg, U. Associations between salivary cytokines and periodontal and microbiological parameters in orthodontic pa-tients. Medicine 2021, 100, e24924. [Google Scholar] [CrossRef]
- Kubota, K. CARD-FISH for environmental microorganisms: Technical advancement and future applications. Microbes Environ. 2013, 28, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Hepp, C.; Shiaelis, N.; Robb, N.C.; Vaughan, A.; Matthews, P.C.; Stoesser, N.; Crook, D.; Kapanidis, A.N. Viral detection and identification in 20 min by rapid single-particle fluorescence in-situ hybridization of viral RNA. Sci. Rep. 2021, 11, 19579. [Google Scholar] [CrossRef] [PubMed]
- Mosaddad, S.A.; Tahmasebi, E.; Yazdanian, A.; Rezvani, M.B.; Seifalian, A.; Yazdanian, M.; Tebyanian, H. Oral microbial biofilms: An update. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2005–2019. [Google Scholar] [CrossRef] [PubMed]
- Dige, I.; Nilsson, H.; Kilian, M.; Nyvad, B. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning mi-croscopy and fluorescence in situ hybridization. Eur. J. Oral Sci. 2007, 115, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, M.; Lucchese, A.; Contaldo, M.; Tampoia, M.; Frassanito, M.A.; Lauritano, D.; della Vella, F. ELISA detection of anti-desmoglein 1 and anti-desmoglein 3 and indirect immunofluorescence in oral pemphigus: A retrospective study. Oral Dis. 2021, 28, 1149–1156. [Google Scholar] [CrossRef]
- Wade, W.; Prosdocimi, E. Profiling of Oral Bacterial Communities. J. Dent. Res. 2020, 99, 621–629. [Google Scholar] [CrossRef]
- Salipante, S.J.; Jerome, K.R. Digital PCR—An Emerging Technology with Broad Applications in Microbiology. Clin. Chem. 2019, 66, 117–123. [Google Scholar] [CrossRef]
- Lochman, J.; Zapletalova, M.; Poskerova, H.; Holla, L.I.; Linhartova, P.B. Rapid Multiplex Real-Time PCR Method for the Detection and Quantification of Selected Cariogenic and Periodontal Bacteria. Diagnostics 2019, 10, 8. [Google Scholar] [CrossRef] [Green Version]
- Kuypers, J.; Jerome, K.R. Applications of Digital PCR for Clinical Microbiology. J. Clin. Microbiol. 2017, 55, 1621–1628. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Yang, L.; Paster, B.J.; Ganly, I.; Morris, L.; Pei, Z.; Hayes, R.B. Oral Microbiome Profiles: 16S rRNA Pyrosequencing and Microarray Assay Comparison. PLoS ONE 2011, 6, e22788. [Google Scholar] [CrossRef] [Green Version]
- Mougeot, J.L.; Stevens, C.B.; Cotton, S.L.; Morton, D.S.; Krishnan, K.; Brennan, M.T.; Lockhart, P.B.; Paster, B.J.; Bahrani Mougeot, F.K. Concordance of HOMIM and HOMINGS technologies in the microbiome analysis of clinical samples. J. Oral Microbiol. 2016, 8, 30379. [Google Scholar] [CrossRef] [Green Version]
- Caselli, E.; Fabbri, C.; D’Accolti, M.; Soffritti, I.; Bassi, C.; Mazzacane, S.; Franchi, M. Defining the oral microbiome by whole-genome sequencing and resistome analysis: The complexity of the healthy picture. BMC Microbiol. 2020, 20, 120. [Google Scholar] [CrossRef]
- Fuks, G.; Elgart, M.; Amir, A.; Zeisel, A.; Turnbaugh, P.J.; Soen, Y.; Shental, N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 2018, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Belstrøm, D.; Grande, M.A.; Sembler-Møller, M.L.; Kirkby, N.; Cotton, S.L.; Paster, B.J.; Holmstrup, P. Influence of periodontal treatment on subgingival and salivary microbiotas. J. Periodontol. 2018, 89, 531–539. [Google Scholar] [CrossRef] [Green Version]
- Belstrøm, D.; Paster, B.J.; Fiehn, N.-E.; Bardow, A.; Holmstrup, P. Salivary bacterial fingerprints of established oral disease revealed by the Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS) technique. J. Oral Microbiol. 2016, 8, 30170. [Google Scholar] [CrossRef] [Green Version]
- The Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Sano, H.; Wakui, A.; Kawachi, M.; Washio, J.; Abiko, Y.; Mayanagi, G.; Yamaki, K.; Tanaka, K.; Takahashi, N.; Sato, T. Profiling system of oral microbiota utilizing polymerase chain reaction-restriction fragment length polymor-phism analysis. J. Oral Biosci. 2021, 63, 292–297. [Google Scholar] [CrossRef]
- Takeshita, T.; Nakano, Y.; Yamashita, Y. Improved accuracy in terminal restriction fragment length polymorphism phy-logenetic analysis using a novel internal size standard definition. Oral Microbiol. Immunol. 2007, 22, 419–428. [Google Scholar] [CrossRef]
- Zangeneh, Z.; Abdi-Ali, A.; Khamooshian, K.; Alvandi, A.; Abiri, R. Bacterial variation in the oral microbiota in multiple sclerosis patients. PLoS ONE 2021, 16, e0260384. [Google Scholar] [CrossRef]
- Sun, F.; Ahmed, A.; Wang, L.; Dong, M.; Niu, W. Comparison of oral microbiota in orthodontic patients and healthy individuals. Microb. Pathog. 2018, 123, 473–477. [Google Scholar] [CrossRef]
- Wei, Y.-S.; Chang, Y.-R.; Tsai, Y.-T.; Yang, Y.-T.; Weng, S.-H.; Tseng, L.-F.; Chou, H.-C.; Hu, A.T.; Liao, E.-C.; Chen, H.-Y.; et al. The distribution of cultivable oral anaerobic microbiota identified by MALDI-TOF MS in healthy subjects and in patients with periodontal disease. J. Pharm. Biomed. Anal. 2020, 192, 113647. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-F.; Hou, X.; Xiao, M.; Zhang, L.; Cheng, J.-W.; Zhou, M.-L.; Huang, J.-J.; Zhang, J.-J.; Xu, Y.-C.; Hsueh, P.-R. Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis for the Identification of Pathogenic Microorganisms: A Review. Microorganisms 2021, 9, 1536. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, A.; Martinelli, M.; Motta, F.; Larini, S.; Arcangeletti, M.C.; Medici, M.C.; Chezzi, C.; De Conto, F. Comparison of peptide nucleic acid fluorescence in situ hybridization assays with culture-based matrix-assisted laser desorption/ionization-time of flight mass spectrometry for the identification of bacteria and yeasts from blood cultures and cerebrospinal fluid cultures. Clin. Microbiol. Infect. 2014, 20, O468–O475. [Google Scholar] [PubMed] [Green Version]
- Garg, K.; Meriläinen, L.; Franz, O.; Pirttinen, H.; Quevedo-Diaz, M.; Croucher, S.; Gilbert, L. RETRACTED ARTICLE: Evaluating polymicrobial immune responses in patients suffering from tick-borne diseases. Sci. Rep. 2018, 8, 15932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Qiu, J.; Guo, L.; Li, D.; Xu, D.; Liu, Q. Development overview of Raman-activated cell sorting devoted to bacterial detection at single-cell level. Appl. Microbiol. Biotechnol. 2021, 105, 1315–1331. [Google Scholar] [CrossRef]
- Lee, K.S.; Pereira, F.C.; Palatinszky, M.; Behrendt, L.; Alcolombri, U.; Berry, D.; Wagner, M.; Stocker, R. Optofluidic Raman-activated cell sorting for targeted genome retrieval or cultivation of microbial cells with specific functions. Nat. Protoc. 2020, 16, 634–676. [Google Scholar] [CrossRef]
- Moter, A.; Göbel, U.B. Fluorescence in situ hybridization (FISH) for direct visualization of microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef]
- Valm, A.M.; Welch, J.L.M.; Rieken, C.W.; Hasegawa, Y.; Sogin, M.L.; Oldenbourg, R.; Dewhirst, F.E.; Borisy, G.G. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl. Acad. Sci. USA 2011, 108, 4152–4157. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Shi, Q.; Grodner, B.; Lenz, J.S.; Zipfel, W.R.; Brito, I.L.; De Vlaminck, I. Highly multiplexed spatial mapping of microbial communities. Nature 2020, 588, 676–681. [Google Scholar] [CrossRef]
- Petrich, A.; Rojas, P.; Schulze, J.; Loddenkemper, C.; Giacani, L.; Schneider, T.; Hertel, M.; Kikhney, J.; Moter, A. Fluorescence in situ hybridization for the identification of Treponema pallidum in tissue sections. Int. J. Med Microbiol. 2015, 305, 709–718. [Google Scholar] [CrossRef]
- Zheng, S.W.; Xu, P.; Cai, L.T.; Tan, Z.W.; Guo, Y.T.; Zhu, R.X.; He, Y. The presence of Prevotella melaninogenica within tissue and preliminary study on its role in the pathogenesis of oral lichen planus. Oral Dis. 2021, 28, 1580–1590. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Z.; Tang, N.; Zhao, Y.; Xu, J.; Li, L.; Qian, L.; Zhang, J.; Fan, Y. Microbial Community Analysis of Saliva and Biopsies in Patients with Oral Lichen Planus. Front. Microbiol. 2020, 11, 629. [Google Scholar] [CrossRef]
- Bernardi, S.; Continenza, M.A.; Al-Ahmad, A.; Karygianni, L.; Follo, M.; Filippi, A.; Macchiarelli, G. Streptococcus spp. and Fusobacterium nucleatum in tongue dorsum biofilm from halitosis patients: A fluo-rescence in situ hybridization (FISH) and confocal laser scanning microscopy (CLSM) study. New Microbiol. 2019, 42, 108–113. [Google Scholar] [PubMed]
- Gmür, R.; Lüthi-Schaller, H. A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. J. Microbiol. Methods 2007, 69, 402–405. [Google Scholar] [CrossRef]
- Karygianni, L.; Hellwig, E.; Al-Ahmad, A. Multiplex fluorescence in situ hybridization (M-FISH) and confocal laser scan-ning microscopy (CLSM) to analyze multispecies oral biofilms. Methods Mol. Biol. 2014, 1147, 65–72. [Google Scholar]
- Do Cabo Fernandes, C.; Rechenberg, D.K.; Zehnder, M.; Belibasakis, G.N. Identification of Synergistetes in endodontic infections. Microb. Pathog. 2014, 73, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Bertl, K.; Zijnge, V.; Zatorska, B.; Leonhard, M.; Schneider-Stickler, B.; Harmsen, H.J.M. Oral cavity anaerobic pathogens in biofilm formation on voice prostheses. Head Neck 2014, 37, 524–529. [Google Scholar] [CrossRef] [Green Version]
- Esteves, G.; Pereira, J.; Azevedo, N.; Azevedo, A.; Mendes, L. Friends with Benefits: An Inside Look of Periodontal Microbes’ Interactions Using Fluorescence In Situ Hybridization—Scoping Review. Microorganisms 2021, 9, 1504. [Google Scholar] [CrossRef]
- Hannig, C.; Sorg, J.; Spitzmüller, B.; Hannig, M.; Al-Ahmad, A. Polyphenolic beverages reduce initial bacterial adherence to enamel in situ. J. Dent. 2009, 37, 560–566. [Google Scholar] [CrossRef]
- Hertel, S.; Graffy, L.; Pötschke, S.; Basche, S.; Al-Ahmad, A.; Hoth-Hannig, W.; Hannig, M.; Hannig, C. Effect of Inula viscosa on the pellicle’s protective properties and initial bioadhesion in-situ. Arch. Oral Biol. 2016, 71, 87–96. [Google Scholar] [CrossRef]
- Lyu, X.; Li, C.; Zhang, J.; Wang, L.; Jiang, Q.; Shui, Y.; Chen, L.; Luo, Y.; Xu, X. A Novel Small Molecule, LCG-N25, Inhibits Oral Streptococcal Biofilm. Front. Microbiol. 2021, 12, 654692. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Cheng, X.; Wang, L.; Qiu, W.; Wang, S.; Zhou, Y.; Li, M.; Li, Y.; Cheng, L.; Li, J.; et al. Combinatorial Effects of Arginine and Fluoride on Oral Bacteria. J. Dent. Res. 2014, 94, 344–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Peng, X.; Wang, S.; Han, Q.; Li, B.; Zhou, X.; Ren, B.; Xu, H.H.K.; Weir, M.D.; Li, M.; et al. A novel antibacterial resin-based root canal sealer modified by Dimethylaminododecyl Methacrylate. Sci. Rep. 2019, 9, 10632. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, A.; Karygianni, L.; Wartenhorst, M.S.; Bächle, M.; Hellwig, E.; Follo, M.; Vach, K.; Han, J.S. Bacterial adhesion and biofilm formation on yttria-stabilized, tetragonal zirconia and titanium oral im-plant materials with low surface roughness—An in situ study. J. Med. Microbiol. 2016, 65, 596–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukic, D.; Karygianni, L.; Flury, M.; Attin, T.; Thurnheer, T. Endodontic-Like Oral Biofilms as Models for Multispecies Interactions in Endodontic Diseases. Microorganisms 2020, 8, 674. [Google Scholar] [CrossRef]
- Kommerein, N.; Doll, K.; Stumpp, N.S.; Stiesch, M. Development and characterization of an oral multispecies biofilm implant flow chamber model. PLoS ONE 2018, 13, e0196967. [Google Scholar] [CrossRef] [Green Version]
- Klug, B.; Santigli, E.; Westendorf, C.; Tangl, S.; Wimmer, G.; Grube, M. From Mouth to Model: Combining in vivo and in vitro Oral Biofilm Growth. Front. Microbiol. 2016, 7, 1448. [Google Scholar] [CrossRef]
- Thurnheer, T.; Belibasakis, G.N. Incorporation of staphylococci into titanium-grown biofilms: An in vitro "submucosal" biofilm model for peri-implantitis. Clin. Oral Implants Res. 2016, 27, 890–895. [Google Scholar] [CrossRef] [Green Version]
- Karygianni, L.; Follo, M.; Hellwig, E.; Burghardt, D.; Wolkewitz, M.; Anderson, A.; Al-Ahmad, A. Microscope-Based Imaging Platform for Large-Scale Analysis of Oral Biofilms. Appl. Environ. Microbiol. 2012, 78, 8703–8711. [Google Scholar] [CrossRef] [Green Version]
- Le Bars, P.; Matamoros, S.; Montassier, E.; Le Vacon, F.; Potel, G.; Soueidan, A.; Jordana, F.; De La Cochetière, M.-F. The oral cavity microbiota: Between health, oral disease, and cancers of the aerodigestive tract. Can. J. Microbiol. 2017, 63, 475–492. [Google Scholar] [CrossRef] [Green Version]
- Hooper, S.J.; Crean, S.J.; Fardy, M.J.; Lewis, M.A.; Spratt, D.A.; Wade, W.G.; Wilson, M.J. A molecular analysis of the bacteria present within oral squamous cell carcinoma. J. Med. Microbiol. 2007, 56, 1651–1659. [Google Scholar] [CrossRef] [Green Version]
- Koren, O.; Spor, A.; Felin, J.; Fåk, F.; Stombaugh, J.; Tremaroli, V.; Behre, C.J.; Knight, R.; Fagerberg, B.; Ley, R.E.; et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl. Acad. Sci. USA 2011, 108, 4592–4598. [Google Scholar] [CrossRef] [Green Version]
- Riviere, G.R.; Riviere, K.H.; Smith, K.S. Molecular and immunological evidence of oral Treponema in the human brain and their association with Alzheimer’s disease. Oral Microbiol. Immunol. 2002, 17, 113–118. [Google Scholar] [CrossRef]
- Shao, D.T.; Li, M.J.; Chen, R.; Wei, W.W. Progress in research of influencing factors of oral microbiome and association between oral microbiome and upper gastrointestinal cancer. Chin J Epidemiol 2020, 41, 1160–1164. [Google Scholar]
- Yakun, J.; Junyi, L. Fluorescence in situ hybridization and its application in the detection of oral microorganisms. Int. J. Stomatol. 2006, 41, 12–14. [Google Scholar]
- Sunde, P.T.; Olsen, I.; Göbel, U.B.; Theegarten, D.; Winter, S.; Debelian, G.J.; Tronstad, L.; Moter, A. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymp-tomatic root-filled teeth. Microbiology 2003, 149, 1095–1102. [Google Scholar] [CrossRef] [Green Version]
- Escapa, I.F.; Chen, T.; Huang, Y.; Gajare, P.; Dewhirst, F.E.; Lemon, K.P. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 2018, 3, e00187-18. [Google Scholar] [CrossRef] [Green Version]
- Yujiao, S.; Yong, W.; Xia, H. Application of fluorescence in situ hybridization in analysis of environmental microbial ecology. Tech. Equip. Environ. Pollut. Control. 2004, 5(11), 14–20. [Google Scholar]
- Weerkamp, F.; Dekking, E.; Ng, Y.Y.; Van Der Velden, V.H.J.; Wai, H.; Böttcher, S.; Brüggemann, M.; Van Der Sluijs, A.J.; Koning, A.; Boeckx, N.; et al. Flow cytometric immunobead assay for the detection of BCR–ABL fusion proteins in leukemia patients. Leukemia 2009, 23, 1106–1117. [Google Scholar] [CrossRef]
- Foster, J.S.; Kolenbrander, P.E. Development of a Multispecies Oral Bacterial Community in a Saliva-Conditioned Flow Cell. Appl. Environ. Microbiol. 2004, 70, 4340–4348. [Google Scholar] [CrossRef] [Green Version]
- Mark Welch, J.; Rossetti, B.; Rieken, C.; Dewhirst, F.; Borisky, G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, Y.J.; Hwang, Y.S. Antibacterial and antioxidant effect of ethanol extracts of Terminalia chebula on Streptococcus mutans. Clin. Exp. Dent. Res. 2021, 7, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Sol, A.; Feuerstein, O.; Featherstone, J.D.B.; Steinberg, D. Effect of sublethal CO2 laser irradiation on gene expression of streptococcus mutans immobilized in a biofilm. Caries Res. 2011, 45, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Koo, H.; Ren, D. Effects of Material Properties on Bacterial Adhesion and Biofilm Formation. J. Dent. Res. 2015, 94, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Araujo, I.J.D.S.; Ricardo, M.G.; Gomes, O.P.; Giovani, P.A.; Puppin-Rontani, J.; Pecorari, V.A.; Martinez, E.F.; Napimoga, M.H.; Nociti Junior, F.H. Titanium dioxide nanotubes added to glass ionomer cements affect S. mutans viability and mechanisms of viru-lence. Braz. Oral Res. 2021, 35, e062. [Google Scholar] [CrossRef]
- Rajapaksha, P.; Elbourne, A.J.; Gangadoo, S.; Brown, R.; Cozzolino, D.; Chapman, J. A review of methods for the detection of pathogenic microorganisms. Analyst 2018, 144, 396–411. [Google Scholar] [CrossRef]
- Jackson, R.; Maarsingh, J.D.; Herbst-Kralovetz, M.M.; Van Doorslaer, K. 3D Oral and Cervical Tissue Models for Studying Papillomavirus Host-Pathogen Interactions. Curr. Protoc. Microbiol. 2020, 59, e129. [Google Scholar] [CrossRef]
- Marx, V. Engineers embrace microbiome messiness. Nat. Chem. Biol. 2019, 16, 581–584. [Google Scholar] [CrossRef]

| Method | Resolution | Culture Reliance | Unsearched Species | Quantification | Microbiota Analysis | Oral Microbiology Applications | Specialty | Reference |
|---|---|---|---|---|---|---|---|---|
| Fluorescence in situ hybridization (FISH) | Genus to Species | No | Detectable | Semi-quantification (affected by hybridizing rate) | Digital image analysis provides spatial resolution | Experimental study Rapid Clinical detection and diagnosis | Intuitive imaging and spectral quantitative positioning analysis | [13,33,34,36] |
| Polymerase chain reaction (PCR) | Subspecies to strain | Isolate nucleic acids from pure cultured bacterial cells | Detectable | Absolute quantification without calibrated standard or highly efficient amplification | Detect and quantify the community compositions | Experimental study Clinical detection and diagnosis | Some subtypes can identify microbes by themselves or be as a sample preparing process for other identification methods | [66,67,68,69] |
| DNA Microarray | Species | No | Only detectable to previously identified species | Quantitative detection of multiple bacteria | Monitor the changes of multiple species, capture the major species | Experimental study Epidemiologic investigation | Based on the pre-constructed microarray chip | [70,71] |
| Next-generation sequencing of 16s rRNA | Subspecies | No | Detectable | Unable | Assess taxonomic diversity of microbiota | Experimental study Clinical treatment evaluation Develop the databases of bacterial genomes | Mainly applied for community composition, evolutionary relationships, and diversity | [71,72,73,74] |
| Next-generation sequencing of whole-genome sequencing | Strain | No | Detectable | Absolute quantification | Unable | Develop the databases of bacterial genomes | Massive DNA sequencing with a high throughput but high cost | [73,75,76] |
| Restriction fragment length polymorphism (RFLP) | Species of several genera | Same as PCR | Detectable | Unable | Assess the diversity of complex microbiota and rapidly compare the structure from different environments | Oral microbiota analysis in smaller laboratories | An inexpensive but complex identification via obtained RFLP patterns | [77,78] |
| Denaturing gradient gel electrophoresis (DGGE) | Species | No, based on 16S rDNA amplified by PCR | Detectable | Semi-quantitative accompanied by real-time PCR | Generate 16S rDNA band patterns as species fingerprints | Experimental study Clinical treatment evaluation | isolate at least 10 different bacteria in each sample | [79,80] |
| Matrix-assisted laser desorption/ionization-Time-of-flight mass spectrometry (MALDI-TOF MS) | Genus to Species | Directly identified by protein or nucleic acid from samples, but the accuracy is higher after separation and purification | detectable | Relative quantification of targeted biomarkers | Mass spectral patterns represent bacterial distribution and relative abundance | Experimental study Rapid clinical detection and diagnosis | Significantly decreases the processing time, but requires expensive instrumentation fail in the identification of mixed infections | [81,82,83] |
| Enzyme-linked immunosorbent assay (ELISA) | Species | No | Only detectable to targeted species corresponding to infection | The level of inflammatory cytokines and immunoglobulins correlate to bacterial load | Unable | Clinical infection diagnostic examinations | Commonly adjunctive tool in clinical practice | [60,65,84] |
| Single-cell Raman spectra (SCRS) | Species | No | Detectable | Quantitative detection of individual droplets | Explore the mechanism of individual microorganisms, but unable to discern bacteria in complex environments | Single-cell investigation Rapid identification and classification | Label-free and non-destructive, but low throughput | [58,85,86] |
| Areas | Aspects | Ref | ||
|---|---|---|---|---|
| Diagnosis assistance | Etiological detection | Human immunodeficiency virus | [90] | |
| Epstein-Barr virus | ||||
| Influenza virus | ||||
| Avian infectious bronchitis virus | ||||
| SARS-CoV-2 | ||||
| Treponema pallidum | ||||
| Target therapy | Oral lichen planus | Prevotella melaninogenica | [91,92] | |
| Capnocytophaga | ||||
| Gemella | ||||
| Escherichia-Shigella | ||||
| Megasphaera | ||||
| Carnobacteriaceae | ||||
| Flavobacteriaceae | ||||
| Halitosis | Eubacteria | [93] | ||
| Fusobacterium nucleatum | ||||
| Streptococcus spp. | ||||
| Oral microecology | Oral microflora identification | In situ identification | [93,94] | |
| Biomass quantification | [95] | |||
| 3D spatial distribution | [89,94] | |||
| Cobacteria in infection | [96] | |||
| Community monitoring | Process of invasion, colonization and transmission | [59,97] | ||
| Interactions between colonies | [98] | |||
| Microbial identity & location map | [89] | |||
| Effects of external factors | [15] | |||
| Occurrence and progression of the disease | [98] | |||
| Effectiveness evaluation | Molecules | Natural molecules | Polyphenol beverages: reduce the adhesion of initial bacteria | [99] |
| Inula viscosa: anti-acid and initial biofilm formation effect | [100] | |||
| Artificial compounds | LCG-N25: adjuvant for the treatment of caries | [101] | ||
| Combined molecules | Arginine + fluoride: synergistic control of dental caries | [102] | ||
| DMADDM + EndoREZ: clinical treatment of periapical periodontitis | [103] | |||
| Oral materials | Dental implant materials | Bacteriostatic effect: oral streptococcus content in the biofilm | [104] | |
| The establishment of models and methods | Oral biofilm model of dental pulp disease established on hydroxyapatite and dentin disc | [105] | ||
| Repeatable and easy-to-use model for cultivating oral multi-species biofilms in a flow chamber system | [106] | |||
| In vivo and in vitro oral biofilm growth model | [107] | |||
| In vitro “submucosal” biofilm model for peri-implantitis | [108] | |||
| FISH: a microscopic method for macroscopic non-invasive monitoring of oral biofilms | [109] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, J.; Wang, H.; Zhang, M.; Xiong, Y.; Yang, L.; Ren, B.; Huang, R. Application of Fluorescence In Situ Hybridization (FISH) in Oral Microbial Detection. Pathogens 2022, 11, 1450. https://doi.org/10.3390/pathogens11121450
Gu J, Wang H, Zhang M, Xiong Y, Yang L, Ren B, Huang R. Application of Fluorescence In Situ Hybridization (FISH) in Oral Microbial Detection. Pathogens. 2022; 11(12):1450. https://doi.org/10.3390/pathogens11121450
Chicago/Turabian StyleGu, Junjie, Huayu Wang, Mengye Zhang, Yichen Xiong, Lei Yang, Biao Ren, and Ruijie Huang. 2022. "Application of Fluorescence In Situ Hybridization (FISH) in Oral Microbial Detection" Pathogens 11, no. 12: 1450. https://doi.org/10.3390/pathogens11121450
APA StyleGu, J., Wang, H., Zhang, M., Xiong, Y., Yang, L., Ren, B., & Huang, R. (2022). Application of Fluorescence In Situ Hybridization (FISH) in Oral Microbial Detection. Pathogens, 11(12), 1450. https://doi.org/10.3390/pathogens11121450