Tracing the Genetic Evolution of Canine Parvovirus Type 2 (CPV-2) in Thailand
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. CPV-2 Genomic DNA Extraction, VP2 Gene Amplification, Sanger Sequencing and Analysis
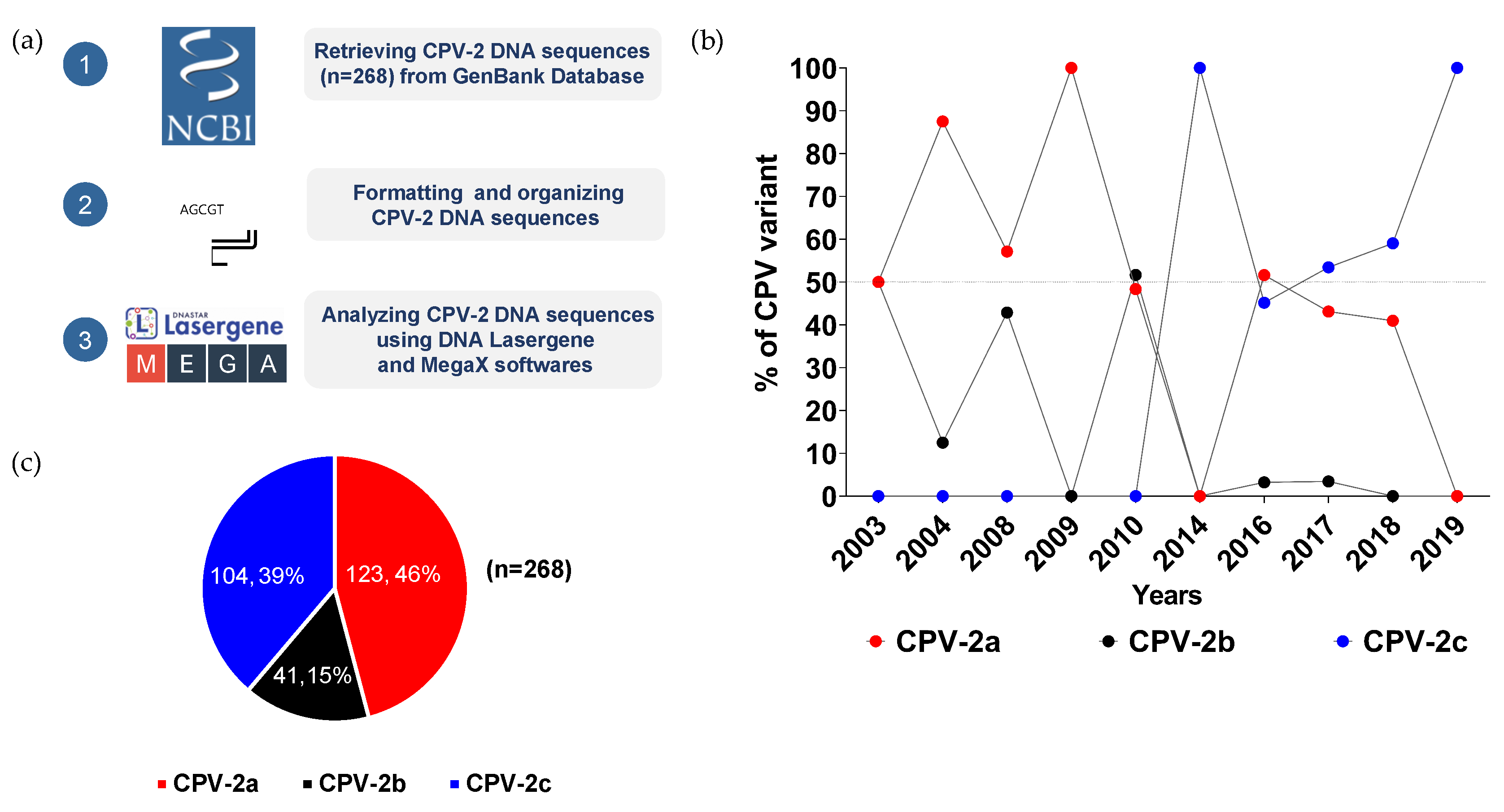
2.3. CPV-2 Sequence Retrieval
2.4. Data Analysis and Phylogenetic Tree Construction
3. Results
3.1. Dynamic changes of CPV-2 in Thailand between 2003 and 2019
3.2. Genomic Evolution of CPV-2a Variant
3.3. Genetic Alteration of CPV-2b Variant
3.4. The Emergence of CPV-2c Antigenic Variant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mazzaferro, E.M. Update on Canine Parvoviral Enteritis. Vet. Clin. N. Am. Small Anim. Pract. 2020, 50, 1307–1325. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Buonavoglia, C. Canine Parvovirus—A Review of Epidemiological and Diagnostic Aspects, with Emphasis on Type 2c. Vet. Microbiol. 2012, 155, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Buonavoglia, C.; Barrs, V.R. Canine Parvovirus Vaccination and Immunisation Failures: Are We Far from Disease Eradication? Vet. Microbiol. 2020, 247, 108760. [Google Scholar] [CrossRef] [PubMed]
- Horecka, K.; Porter, S.; Amirian, E.S.; Jefferson, E. A Decade of Treatment of Canine Parvovirus in an Animal Shelter: A Retrospective Study. Animals 2020, 10, 939. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.-M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef]
- TRUYEN, U.; EVERMANN, J.F.; VIELER, E.; PARRISH, C.R. Evolution of Canine Parvovirus Involved Loss and Gain of Feline Host Range. Virology 1996, 215, 186–189. [Google Scholar] [CrossRef] [Green Version]
- Parrish, C.R. 3 Pathogenesis of Feline Panleukopenia Virus and Canine Parvovirus. Baillière’s Clin. Haematol. 1995, 8, 57–71. [Google Scholar] [CrossRef]
- Hoelzer, K.; Shackelton, L.A.; Parrish, C.R.; Holmes, E.C. Phylogenetic Analysis Reveals the Emergence, Evolution and Dispersal of Carnivore Parvoviruses. J. Gen. Virol. 2008, 89, 2280–2289. [Google Scholar] [CrossRef]
- Reed, A.P.; Jones, E.V.; Miller, T.J. Nucleotide Sequence and Genome Organization of Canine Parvovirus. J. Virol. 1988, 62, 266–276. [Google Scholar] [CrossRef] [Green Version]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High Rate of Viral Evolution Associated with the Emergence of Carnivore Parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef]
- Zhou, P.; Zeng, W.; Zhang, X.; Li, S. The Genetic Evolution of Canine Parvovirus–A New Perspective. PLoS ONE 2017, 12, e0175035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, S.F.; Sgro, J.Y.; Parrish, C.R. Multiple Amino Acids in the Capsid Structure of Canine Parvovirus Coordinately Determine the Canine Host Range and Specific Antigenic and Hemagglutination Properties. J. Virol. 1992, 66, 6858–6867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decaro, N.; Desario, C.; Elia, G.; Martella, V.; Mari, V.; Lavazza, A.; Nardi, M.; Buonavoglia, C. Evidence for Immunisation Failure in Vaccinated Adult Dogs Infected with Canine Parvovirus Type 2c. New Microbiol. 2008, 31, 125–130. [Google Scholar] [PubMed]
- Wang, D.; Yuan, W.; Davis, I.; Parrish, C.R. Nonstructural Protein-2 and the Replication of Canine Parvovirus. Virology 1998, 240, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Stucker, K.M.; Pagan, I.; Cifuente, J.O.; Kaelber, J.T.; Lillie, T.D.; Hafenstein, S.; Holmes, E.C.; Parrish, C.R. The Role of Evolutionary Intermediates in the Host Adaptation of Canine Parvovirus. J. Virol. 2011, 86, 1514–1521. [Google Scholar] [CrossRef] [Green Version]
- Parrish, C.R.; O’Connell, P.H.; Evermann, J.F.; Carmichael, L.E. Natural Variation of Canine Parvovirus. Science 1985, 230, 1046–1048. [Google Scholar] [CrossRef]
- Buonavoglia, C.; Martella, V.; Pratelli, A.; Tempesta, M.; Cavalli, A.; Buonavoglia, D.; Bozzo, G.; Elia, G.; Decaro, N.; Carmichael, L. Evidence for Evolution of Canine Parvovirus Type 2 in Italy. J. Gen. Virol. 2001, 82, 3021–3025. [Google Scholar] [CrossRef]
- Ohshima, T.; Hisaka, M.; Kawakami, K.; Kishi, M.; Tohya, Y.; Mochizuki, M. Chronological Analysis of Canine Parvovirus Type 2 Isolates in Japan. J. Vet. Med. Sci. 2008, 70, 769–775. [Google Scholar] [CrossRef] [Green Version]
- Hao, X.; Li, Y.; Xiao, X.; Chen, B.; Zhou, P.; Li, S. The Changes in Canine Parvovirus Variants over the Years. Int. J. Mol. Sci. 2022, 23, 11540. [Google Scholar] [CrossRef]
- Parrish, C.R.; Have, P.; Foreyt, W.J.; Evermann, J.F.; Senda, M.; Carmichael, L.E. The Global Spread and Replacement of Canine Parvovirus Strains. J. Gen. Virol. 1988, 69, 1111–1116. [Google Scholar] [CrossRef]
- Takano, T.; Hamaguchi, S.; Hasegawa, N.; Doki, T.; Soma, T. Predominance of Canine Parvovirus 2b in Japan: An Epidemiological Study during 2014–2019. Arch. Virol. 2021, 166, 3151–3156. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.; Lin, W.-H.; Le, V.P.; Nga, B.T.T.; Chiou, M.-T.; Lin, C.-N. Molecular Epidemiology of Canine Parvovirus Type 2 in Vietnam from November 2016 to February 2018. Virol. J. 2019, 16, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doan, H.T.T.; Le, X.T.K.; Do, R.T.; Nguyen, K.T.; Le, T.H. Canine Parvovirus Type 2c in Vietnam Continues to Produce Distinct Descendants with New Mutations Restricted to Vietnamese Variants. Arch. Virol. 2021, 166, 1741–1749. [Google Scholar] [CrossRef] [PubMed]
- Ohneiser, S.A.; Hills, S.F.; Cave, N.J.; Passmore, D.; Dunowska, M. Canine Parvoviruses in New Zealand Form a Monophyletic Group Distinct from the Viruses Circulating in Other Parts of the World. Vet. Microbiol. 2015, 178, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Kwan, E.; Carrai, M.; Lanave, G.; Hill, J.; Parry, K.; Kelman, M.; Meers, J.; Decaro, N.; Beatty, J.A.; Martella, V.; et al. Analysis of Canine Parvoviruses Circulating in Australia Reveals Predominance of Variant 2b and Identifies Feline Parvovirus-like Mutations in the Capsid Proteins. Transbound. Emerg. Dis. 2021, 68, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Carrino, M.; Tassoni, L.; Campalto, M.; Cavicchio, L.; Mion, M.; Corrò, M.; Natale, A.; Beato, M.S. Molecular Investigation of Recent Canine Parvovirus-2 (CPV-2) in Italy Revealed Distinct Clustering. Viruses 2022, 14, 917. [Google Scholar] [CrossRef] [PubMed]
- Giraldo-Ramirez, S.; Rendon-Marin, S.; Ruiz-Saenz, J. Phylogenetic, Evolutionary and Structural Analysis of Canine Parvovirus (CPV-2) Antigenic Variants Circulating in Colombia. Viruses 2020, 12, 500. [Google Scholar] [CrossRef]
- Qi, S.; Zhao, J.; Guo, D.; Sun, D. A Mini-Review on the Epidemiology of Canine Parvovirus in China. Front. Vet. Sci. 2020, 7, 5. [Google Scholar] [CrossRef]
- Castillo, C.; Neira, V.; Aniñir, P.; Grecco, S.; Pérez, R.; Panzera, Y.; Zegpi, N.-A.; Sandoval, A.; Sandoval, D.; Cofre, S.; et al. First Molecular Identification of Canine Parvovirus Type 2 (CPV2) in Chile Reveals High Occurrence of CPV2c Antigenic Variant. Front. Vet. Sci. 2020, 7, 194. [Google Scholar] [CrossRef]
- Gagnon, C.A.; Allard, V.; Cloutier, G. Canine Parvovirus Type 2b Is the Most Prevalent Genomic Variant Strain Found in Parvovirus Antigen Positive Diarrheic Dog Feces Samples across Canada. Can. Vet. J. La Rev. Vet. Can. 2016, 57, 29–31. [Google Scholar]
- Phromnoi, S.; Sirinarumitr, K.; Sirinarumitr, T. Sequence Analysis of VP2 Gene of Canine Parvovirus Isolates in Thailand. Virus Genes 2010, 41, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Charoenkul, K.; Tangwangvivat, R.; Janetanakit, T.; Boonyapisitsopa, S.; Bunpapong, N.; Chaiyawong, S.; Amonsin, A. Emergence of Canine Parvovirus Type 2c in Domestic Dogs and Cats from Thailand. Transbound. Emerg. Dis. 2019, 66, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Inthong, N.; Kaewmongkol, S.; Meekhanon, N.; Sirinarumitr, K.; Sirinarumitr, T. Dynamic Evolution of Canine Parvovirus in Thailand. Vet. World 2020, 13, 245–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Torre, D.; Mafla, E.; Puga, B.; Erazo, L.; Astolfi-Ferreira, C.; Ferreira, A.P. Molecular Characterization of Canine Parvovirus Variants (CPV-2a, CPV-2b, and CPV-2c) Based on the VP2 Gene in Affected Domestic Dogs in Ecuador. Vet. World 2018, 11, 480–487. [Google Scholar] [CrossRef] [Green Version]
- Chiang, S.-Y.; Wu, H.-Y.; Chiou, M.-T.; Chang, M.-C.; Lin, C.-N. Identification of a Novel Canine Parvovirus Type 2c in Taiwan. Virol. J. 2016, 13, 160. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wang, J.; Jiang, Y.; Cheng, Y.; Lin, P.; Zhu, H.; Han, G.; Yi, L.; Zhang, S.; Guo, L.; et al. Typing of Canine Parvovirus Strains Circulating in North-East China. Transbound. Emerg. Dis. 2017, 64, 495–503. [Google Scholar] [CrossRef]
- Balboni, A.; Niculae, M.; Vito, S.D.; Urbani, L.; Terrusi, A.; Muresan, C.; Battilani, M. The Detection of Canine Parvovirus Type 2c of Asian Origin in Dogs in Romania Evidenced Its Progressive Worldwide Diffusion. BMC Vet. Res. 2021, 17, 206. [Google Scholar] [CrossRef]
- Hong, C.; Decaro, N.; Desario, C.; Tanner, P.; Pardo, M.C.; Sanchez, S.; Buonavoglia, C.; Saliki, J.T. Occurrence of Canine Parvovirus Type 2c in the United States. J. Vet. Diagn. Investig. 2007, 19, 535–539. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Guo, D.; Li, C.; Wang, E.; Wei, S.; Wang, Z.; Yao, S.; Zhao, X.; Su, M.; Wang, X.; et al. Co-Circulation of the Rare CPV-2c with Unique Gln370Arg Substitution, New CPV-2b with Unique Thr440Ala Substitution, and New CPV-2a with High Prevalence and Variation in Heilongjiang Province, Northeast China. PLoS ONE 2015, 10, e0137288. [Google Scholar] [CrossRef]
- Sanjuán, R.; Nebot, M.R.; Chirico, N.; Mansky, L.M.; Belshaw, R. Viral Mutation Rates. J. Virol. 2010, 84, 9733–9748. [Google Scholar] [CrossRef] [Green Version]
- Tsao, J.; Chapman, M.S.; Agbandje, M.; Keller, W.; Smith, K.; Wu, H.; Luo, M.; Smith, T.J.; Rossmann, M.G.; Compans, R.W.; et al. The Three-Dimensional Structure of Canine Parvovirus and Its Functional Implications. Science 1991, 251, 1456–1464. [Google Scholar] [CrossRef] [PubMed]
- Alexis, V.-A.; Sonia, V.; Daniela, S.; Miguel, G.; Timothy, H.; Valentina, F.; Lisette, L.; Leonardo, S. Molecular Analysis of Full-Length VP2 of Canine Parvovirus Reveals Antigenic Drift in CPV-2b and CPV-2c Variants in Central Chile. Animals 2021, 11, 2387. [Google Scholar] [CrossRef] [PubMed]
- Ogbu, K.I.; Mira, F.; Purpari, G.; Nwosuh, C.; Loria, G.R.; Schirò, G.; Chiaramonte, G.; Tion, M.T.; Bella, S.D.; Ventriglia, G.; et al. Nearly Full-length Genome Characterization of Canine Parvovirus Strains Circulating in Nigeria. Transbound. Emerg. Dis. 2020, 67, 635–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndiana, L.A.; Lanave, G.; Zarea, A.A.K.; Desario, C.; Odigie, E.A.; Ehab, F.A.; Capozza, P.; Greco, G.; Buonavoglia, C.; Decaro, N. Molecular Characterization of Carnivore Protoparvovirus 1 Circulating in Domestic Carnivores in Egypt. Front. Vet. Sci. 2022, 9, 932247. [Google Scholar] [CrossRef] [PubMed]
- Mira, F.; Purpari, G.; Bella, S.D.; Colaianni, M.L.; Schirò, G.; Chiaramonte, G.; Gucciardi, F.; Pisano, P.; Lastra, A.; Decaro, N.; et al. Spreading of Canine Parvovirus Type 2c Mutants of Asian Origin in Southern Italy. Transbound. Emerg. Dis. 2019, 66, 2297–2304. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jantafong, T.; Ruenphet, S.; Garner, H.R.; Ritthipichai, K. Tracing the Genetic Evolution of Canine Parvovirus Type 2 (CPV-2) in Thailand. Pathogens 2022, 11, 1460. https://doi.org/10.3390/pathogens11121460
Jantafong T, Ruenphet S, Garner HR, Ritthipichai K. Tracing the Genetic Evolution of Canine Parvovirus Type 2 (CPV-2) in Thailand. Pathogens. 2022; 11(12):1460. https://doi.org/10.3390/pathogens11121460
Chicago/Turabian StyleJantafong, Tippawan, Sakchai Ruenphet, Harold R. Garner, and Krit Ritthipichai. 2022. "Tracing the Genetic Evolution of Canine Parvovirus Type 2 (CPV-2) in Thailand" Pathogens 11, no. 12: 1460. https://doi.org/10.3390/pathogens11121460




