Description of Male, Redescription of Female, Host Record, and Phylogenetic Position of Haemaphysalis danieli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
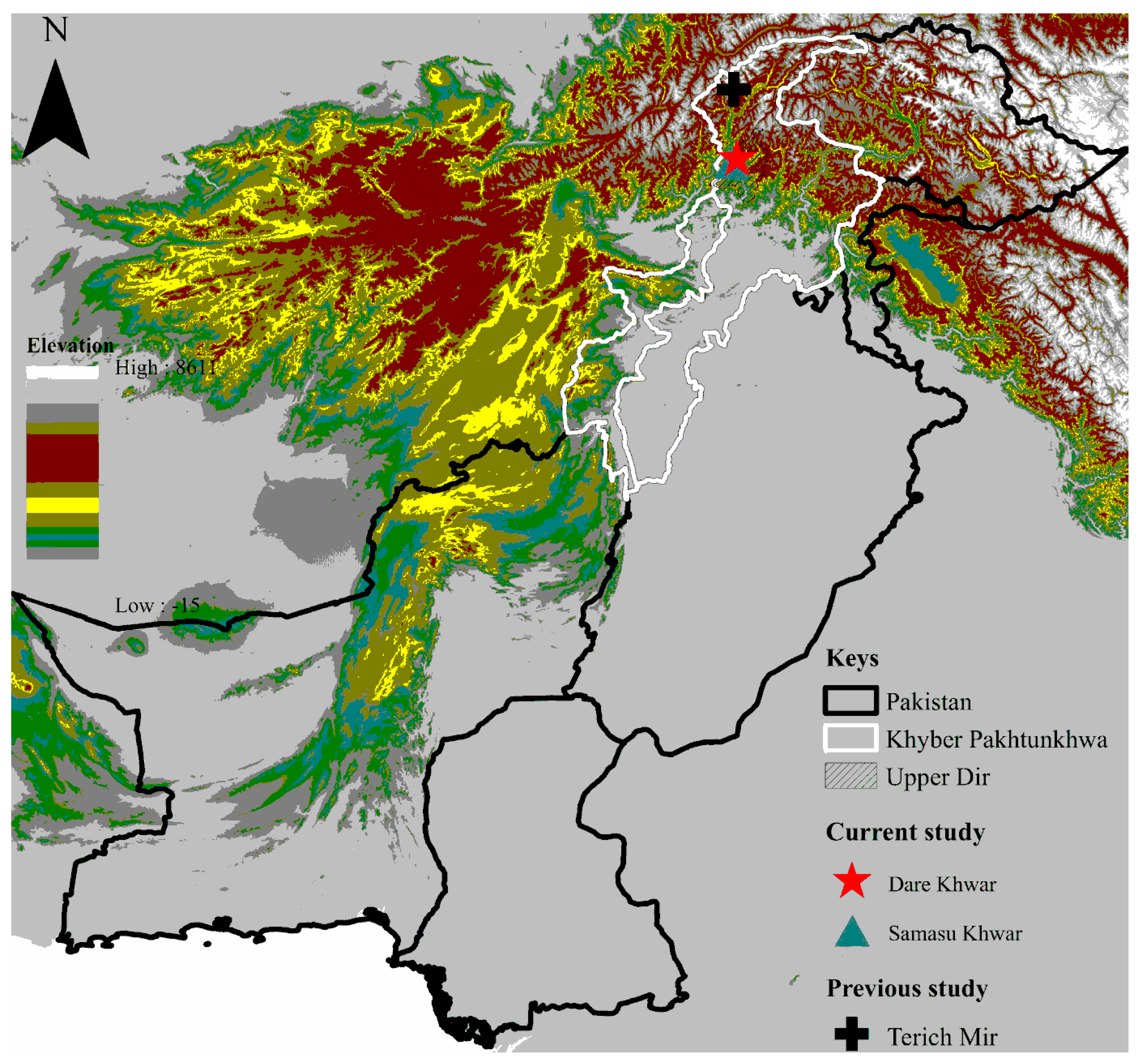
2.2. Study Area
2.3. Tick Collection and Preservation
2.4. Morphological Identification of Ticks
2.5. DNA Extraction and PCR
2.6. DNA Sequencing and Phylogenetic Analyses
3. Results
3.1. Collected Ticks and Host Record
3.2. Description of Haemaphysalis danieli
3.2.1. Description of Male
3.2.2. Redescription of Female
3.3. Molecular Analyses
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Estrada-Peña, A.; Pfäffle, M.P.; Petney, T.N. Genus Haemaphysalis Koch, 1844. In Ticks of Europe and North Africa; Springer: Cham, Switzerland, 2017; pp. 225–229. [Google Scholar]
- Guglielmone, A.A.; Petney, T.N.; Robbins, R.G. Ixodidae (Acari: Ixodoidea): Descriptions and redescriptions of all known species from 1758 to December 31, 2019. Zootaxa 2020, 4871, 1–322. [Google Scholar] [CrossRef]
- Černý, V.; Hoogstraal, H. Haemaphysalis (Allophysalis) danieli, sp. n. (Ixodoidea: Ixodidae), female and tentatively associated immature stages from high mountains of northern Pakistan and Afghanistan. J. Parasitol. Res. 1977, 63, 567–574. [Google Scholar] [CrossRef]
- Hoogstraal, H.; Kim, K.C. Tick and mammal coevolution, with emphasis on Haemaphysalis. In Coevolution of Parasitic Arthropods and Mammals; Kim, K.C., Ed.; Wiley-Inter-Sci.: New York, NY, USA, 1985; Volume 69, pp. 505–568. [Google Scholar]
- Hoogstraal, H. Haemaphysalis tibetensis sp. n. and its significance in elucidating phylogenetic patterns in the genus (Ixodoidea, Ixodidae). J. Parasitol. Res. 1965, 51, 452–459. [Google Scholar] [CrossRef]
- Hoogstraal, H. Haemaphysalis (Allophysalis) pospelovashtromae sp. n. from USSR and redescription of the type of material of H. (A.) warburtoni Nuttall from China (Ixodoidea, Ixodidae). J. Parasitol. Res. 1966, 52, 787–800. [Google Scholar] [CrossRef]
- Dhanda, V.; Bhat, H.R. Haemaphysalis (Allophysalis) garhwalensis sp. n. (Acarina: Ixodidae) parasitizing sheep and goats in Garhwal region, Uttar Pradesh, India. J. Parasitol. Res. 1968, 54, 674–678. [Google Scholar] [CrossRef]
- Hoogstraal, H.; Wassef, H.Y. Haemaphysalis (Allophysalis) kopetdaghica: Identity and discovery of each feeding stage on the wild goat in northern Iran (Ixodoidea: Ixodidae). J. Parasitol. Res. 1979, 65, 783–790. [Google Scholar] [CrossRef]
- Geevarghese, G.; Mishra, A.C. Haemaphysalis Ticks of India; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Sylla, M.; Ndiaye, M.; Souris, M.; Gonzalez, J.P. Ticks (Acari: Ixodida) of the genus Haemaphysalis Koch, 1844 in Senegal: A review of host associations, chorology, and identification. Acarologia 2018, 58, 928–945. [Google Scholar] [CrossRef]
- Orkun, Ö.; Vatansever, Z. Rediscovery and first genetic description of some poorly known tick species: Haemaphysalis kopetdaghica Kerbabaev, 1962 and Dermacentor raskemensis Pomerantzev, 1946. Ticks Tick Borne Dis. 2021, 12, 101726. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A laboratory Manual (No. Ed. 2); Cold Spring Harbor Laboratory Press: Cold Spring, NY, USA, 1989. [Google Scholar]
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA-X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Apanaskevich, D.A.; Goodman, S.M. Description of a new species of Haemaphysalis Koch, 1844 (Acari: Ixodidae) from the H. (Rhipistoma) asiatica subgroup, parasite of an endemic Malagasy carnivoran (Carnivora: Eupleridae). Syst. Parasitol. 2020, 97, 591–599. [Google Scholar] [CrossRef]
- Apanaskevich, D.A.; Tomlinson, J.A. Description of two new species of Haemaphysalis Koch, 1844 (Acari: Ixodidae) from the H. (Rhipistoma) spinulosa subgroup, parasites of carnivores and other mammals in Africa. Syst. Parasitol. 2020, 97, 601–621. [Google Scholar] [CrossRef]
- Kiszewski, A.E.; Matuschka, F.R.; Spielman, A. Mating strategies and spermiogenesis in ixodid ticks. Annu. Rev. Entomol. 2001, 46, 167–182. [Google Scholar] [CrossRef]
- Engelstädter, J.; Hurst, G.D. The ecology and evolution of microbes that manipulate host reproduction. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 127–149. [Google Scholar] [CrossRef]
- Duron, O.; Binetruy, F.; Noël, V.; Cremaschi, J.; McCoy, K.D.; Arnathau, C.; Plantard, O.; Goolsby, J.; Pérez de León, A.A.; Heylen, D.J.; et al. Evolutionary changes in symbiont community structure in ticks. Mol. Ecol. 2017, 26, 2905–2921. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H. Sex ratio adaptations to local mate competition in a parasitic wasp. Science 1980, 208, 1157–1159. [Google Scholar] [CrossRef]
- Peñalver, E.; Arillo, A.; Delclòs, X.; Peris, D.; Grimaldi, D.A.; Anderson, S.R.; Nascimbene, P.C.; Pérez-de la Fuente, R. Ticks parasitised feathered dinosaurs as revealed by Cretaceous amber assemblages. Nat. Commun. 2017, 8, 1924. [Google Scholar] [CrossRef]
- Teng, K.F. On the Chinese Haemaphysalis subgenus Allophysalis with description of a new species. Acta Entomol. Sin. 1980, 23, 86–89. [Google Scholar]
- Guglielmone, A.A.; Robbins, R.G.; Apanaskevich, D.A.; Petney, T.N.; Estrada-Peña, A.; Horak, I.G. The Hard Ticks of the World; Springer: Dordrecht, The Netherlands, 2014; Volume 10, pp. 978–994. [Google Scholar]
- Barker, S.C.; Murrell, A. Systematics and evolution of ticks with a list of valid genus and species names. Parasitology 2004, 129, S15–S36. [Google Scholar] [CrossRef]
- Beati, L.; Klompen, H. Phylogeography of ticks (Acari: Ixodida). Annu. Rev. Entomol. 2018, 64, 379–397. [Google Scholar] [CrossRef]
- Mans, B.J.; Featherston, J.; Kvas, M.; Pillay, K.A.; de Klerk, D.G.; Pienaar, R.; de Castro, M.H.; Schwan, T.G.; Lopez, J.E.; Teel, P.; et al. Argasid and ixodid systematics: Implications for soft tick evolution and systematics, with a new argasid species list. Ticks Tick Borne Dis. 2019, 10, 219–240. [Google Scholar] [CrossRef]
- Thompson, A.T.; Dominguez, K.; Cleveland, C.A.; Dergousoff, S.J.; Doi, K.; Falco, R.C.; Greay, T.; Irwin, P.; Lindsay, L.R.; Liu, J.; et al. Molecular characterization of Haemaphysalis species and a molecular genetic key for the identification of Haemaphysalis of North America. Front. Vet. Sci. 2020, 7, 141. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.; Numan, M.; Khan, M.; Aiman, O.; Muñoz-Leal, S.; Chitimia-Dobler, L.; Labruna, M.B.; Nijhof, A.M. Ornithodoros (Pavlovskyella) ticks associated with a Rickettsia sp. in Pakistan. Parasit. Vectors 2022, 15, 1–13. [Google Scholar] [CrossRef]
- Alam, S.; Khan, M.; Alouffi, A.; Almutairi, M.M.; Ullah, S.; Numan, M.; Islam, N.; Khan, Z.; Aiman, O.; Zaman Safi, S.; et al. Spatio-temporal patterns of ticks and molecular survey of Anaplasma marginale, with notes on their phylogeny. Microorganisms 2022, 10, 1663. [Google Scholar] [CrossRef]
- Burger, T.D.; Shao, R.; Beati, L.; Miller, H.; Barker, S.C. Phylogenetic analysis of ticks (Acari: Ixodida) using mitochondrial genomes and nuclear rRNA genes indicates that the genus Amblyomma is polyphyletic. Mol. Phylogenet. Evol. 2012, 64, 45–55. [Google Scholar] [CrossRef]
- Aiman, O.; Ullah, S.; Chitimia-Dobler, L.; Nijhof, A.M.; Ali, A. First report of Nosomma monstrosum ticks infesting Asian water buffaloes (Bubalus bubalis) in Pakistan. Ticks Tick Borne Dis. 2022, 13, 101899. [Google Scholar] [CrossRef] [PubMed]




| Target Genes | Sequences (5′-3′) | Amplicon Size | PCR Conditions | References |
|---|---|---|---|---|
| 16S rRNA | 16S+1-CCGGTCTGAACTCAGATCAAGT 16S−1-GCTCAATGATTTTTTAAATTGCTG | 460 bp | 95 °C 3 min, 40 × (95 °C 30 s, 55 °C 60 s, 72 °C 1 min), 72 °C 7 min | [13] |
| cox1 | HC02198-TAAACTTCAGGGTGACCAAAAAATCA LCO1490-GGTCAACAAATCATAAAGATATTGG | 710 bp | 95 °C 30 s, 40 × (95 °C 30 s, 48 °C 30 s, 72 °C 1 min), 72 °C 5 min | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, I.; Ullah, S.; Alouffi, A.; Almutairi, M.M.; Khan, M.; Numan, M.; Safi, S.Z.; Chitimia-Dobler, L.; Tanaka, T.; Ali, A. Description of Male, Redescription of Female, Host Record, and Phylogenetic Position of Haemaphysalis danieli. Pathogens 2022, 11, 1495. https://doi.org/10.3390/pathogens11121495
Ahmad I, Ullah S, Alouffi A, Almutairi MM, Khan M, Numan M, Safi SZ, Chitimia-Dobler L, Tanaka T, Ali A. Description of Male, Redescription of Female, Host Record, and Phylogenetic Position of Haemaphysalis danieli. Pathogens. 2022; 11(12):1495. https://doi.org/10.3390/pathogens11121495
Chicago/Turabian StyleAhmad, Iftikhar, Shafi Ullah, Abdulaziz Alouffi, Mashal M. Almutairi, Mehran Khan, Muhammad Numan, Sher Zaman Safi, Lidia Chitimia-Dobler, Tetsuya Tanaka, and Abid Ali. 2022. "Description of Male, Redescription of Female, Host Record, and Phylogenetic Position of Haemaphysalis danieli" Pathogens 11, no. 12: 1495. https://doi.org/10.3390/pathogens11121495












