Surveillance, Diversity and Vegetative Compatibility Groups of Fusarium oxysporum f. sp. vasinfectum Collected in Cotton Fields in Australia (2017 to 2022)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Disease Surveillance
2.2. Sampling and Pathogen Isolation
2.3. DNA Extraction
2.4. PCR Amplification for the TEF1
2.5. Australian Fov Detection
2.6. Californian Fov Race 4 Detection
2.7. VCG Determination
2.8. Reference Isolates of Australian F. oxysporum f. sp. vasinfectum
2.9. Generating Nitrate Non-Utilising (nit) Mutants
2.10. Pairing nit Mutants in VCG Tests
3. Results
3.1. Surveillance
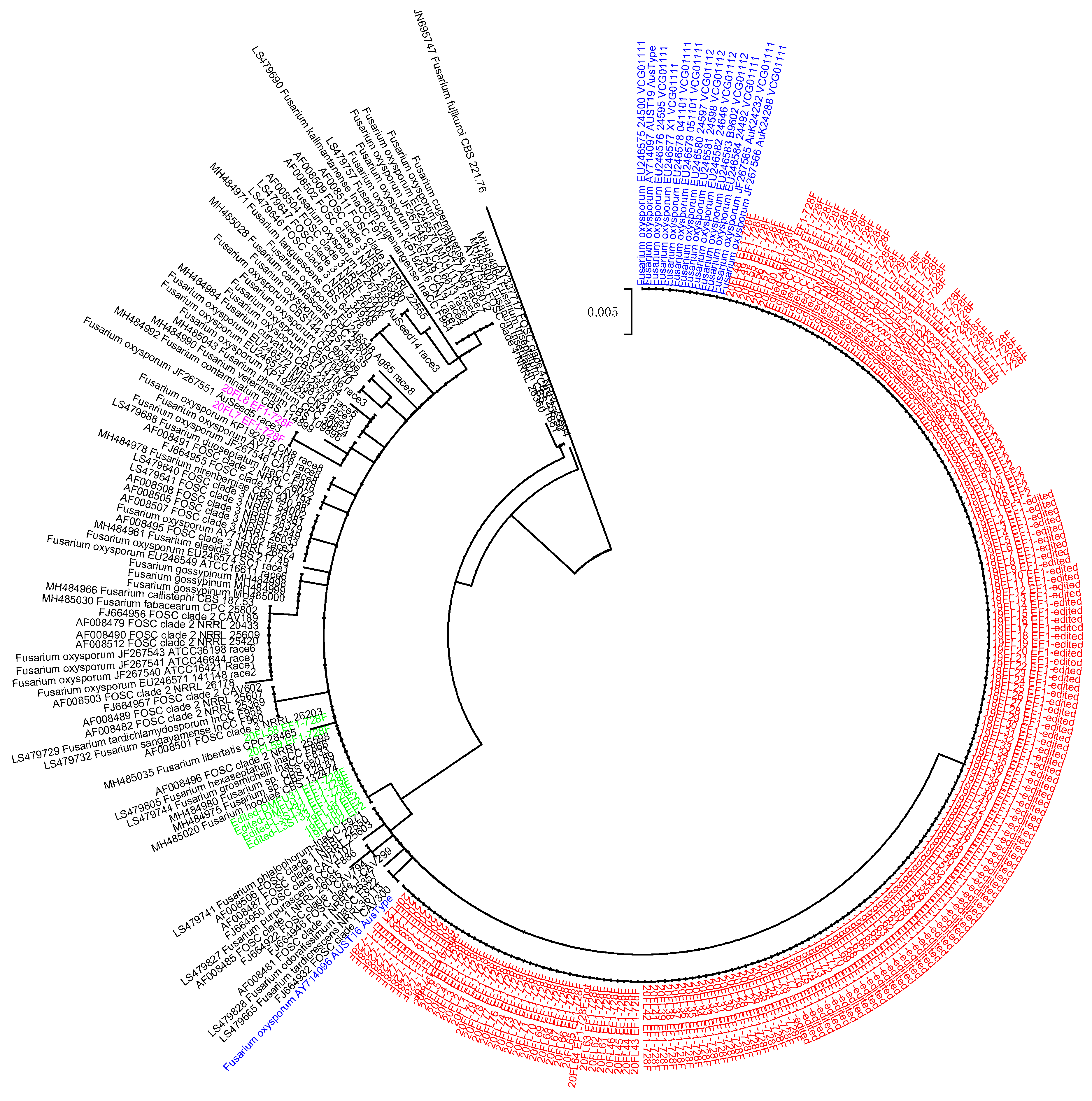
3.2. TEF1-Based Diversity
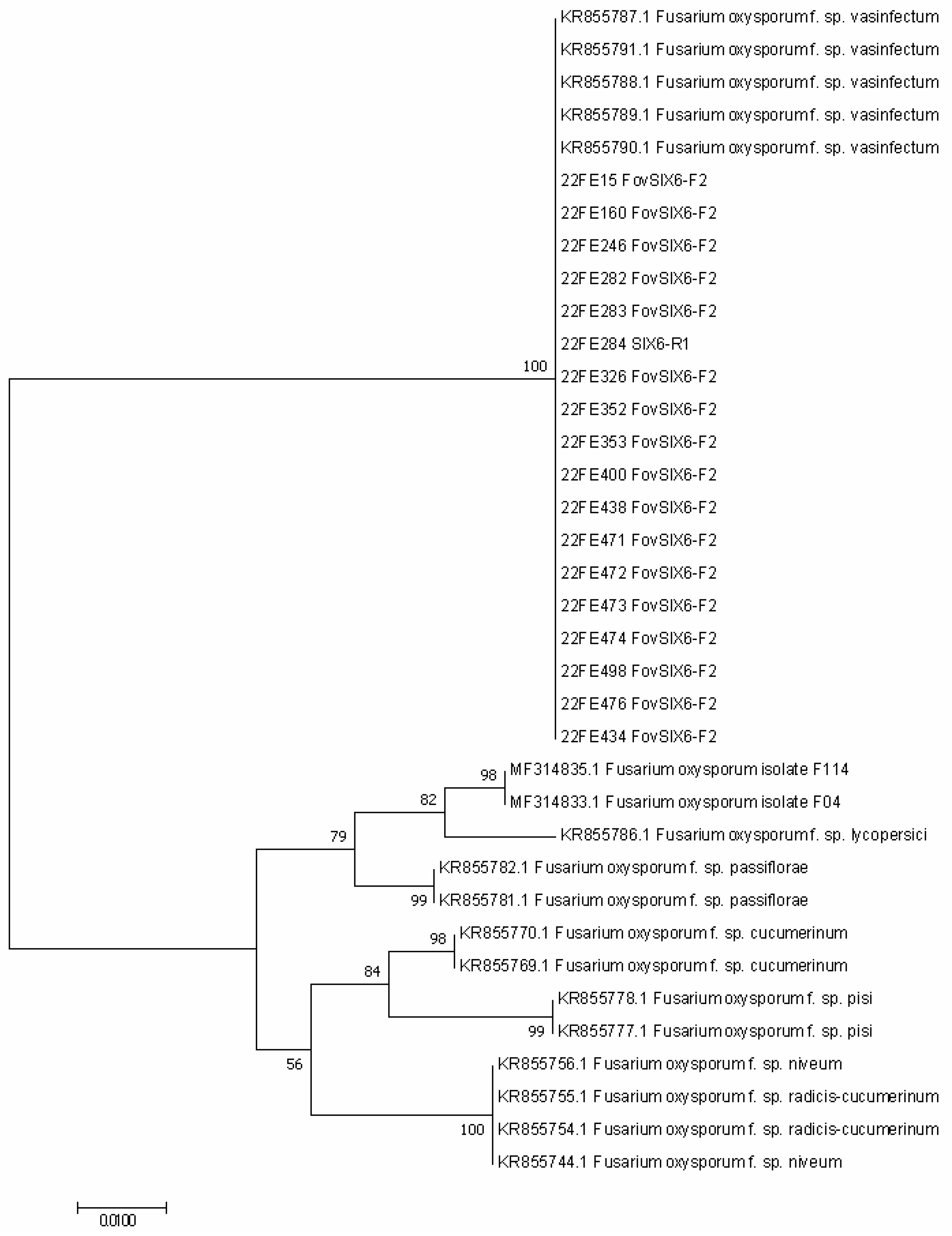
3.3. Australian Fov Identification
3.4. Californian Race 4 Detection
3.5. VCGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- CRDC. CRDC Annual Report 2017–2018; Cotton Research and Development Corporation: Narrabri, Australia, 2018. [Google Scholar]
- Smith, L. Improving the Management of Cotton Diseases in Australian Cotton Farming Systems (RRDP1724); Cotton Research and Development Corporation: Narrabri, Australia, 2020; pp. 1–172. [Google Scholar]
- Kirkby, K.; Lonergan, P.; Allen, S. Three decades of cotton disease surveys in NSW, Australia. Crop Pasture Sci. 2013, 64, 774–779. [Google Scholar] [CrossRef]
- Kochman, J. Fusarium wilt in cotton—A new record in Australia. Australas. Plant Pathol. 1995, 24, 74. [Google Scholar] [CrossRef]
- Bentley, S.; Salmond, G.; O’Neil, W.; Obst, N.; Moore, N.; Kochman, J. Management strategies for Fusarium wilt of cotton. In Proceedings of the 10th Australian Cotton Conference, Brisbane, Australia, 16–18 August 2000; Australian Cotton Growers Association: Wee Waa, Australia, 2000; pp. 455–461. [Google Scholar]
- Allen, S.; Anderson, C.M.T.; Lonergan, P.A.; Scheikowski, L.J.; Smith, L.J. Cotton Pathology Survey 2008/09. In Cotton Pest Management Guide 2009–10; Cotton Catchment Communities CRC: Narrabri, Australia, 2010; pp. 124–127. [Google Scholar]
- CSD. Disease Ranks. 2022. Available online: https://csd.net.au/disease-ranks (accessed on 20 November 2011).
- McCollum, D. Qualitative Report on the 2020–21 Cotton Season: A Survey of Consultants; Cotton Research Development Corporation: Narrabri, Australia, 2021; pp. 1–44. [Google Scholar]
- Davis, R.M.; Colyer, P.D.; Rothrock, C.S.; Kochman, J.K. Fusarium wilt of cotton: Population diversity and implications for management. Plant Dis. 2006, 90, 692–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skovgaard, K.; Nirenberg, H.I.; O’Donnell, K.; Rosendahl, S. Evolution of Fusarium oxysporum f. sp. vasinfectum races inferred from multigene genealogies. Phytopathology 2001, 91, 231–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, R.; Moore, N.; Kochman, J. Characterisation of a population of Fusarium oxysporum f. sp. vasinfectum causing wilt of cotton in Australia. Aust. J. Agric. Res. 1996, 47, 1143–1156. [Google Scholar] [CrossRef]
- Wang, B.; Brubaker, C.L.; Summerell, B.A.; Thrall, P.H.; Burdon, J.J. Local origin of two vegetative compatibility groups of Fusarium oxysporum f. sp. vasinfectum in Australia. Evol. Appl. 2010, 3, 505–524. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Sanogo, S.; Wedegaertner, T.; Nichols, R.; Zhang, J.F. First report of Fusarium solani causing Fusarium wilt in Pima cotton (Gossypium barbadense) in New Mexico, USA. Plant Dis. 2019, 103, 3279. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Sanogo, S.; Wedegaertner, T.; Nichols, R.; Zhang, J.F. First report of cotton (Gossypium) wilt caused by Fusarium proliferatum in New Mexico, USA. Plant Dis. 2019, 103, 2679. [Google Scholar] [CrossRef]
- Zhu, Y.; Abdelraheem, A.; Wedegaertner, T.; Nichols, R.; Zhang, J.F. First report of Fusarium fujikuroi causing wilt on Pima cotton (Gossypium barbadense) seedlings in New Mexico, USA. Plant Dis. 2021, 105, 228. [Google Scholar] [CrossRef]
- O’Donnell, K.; Whitaker, B.K.; Laraba, I.; Proctor, R.H.; Brown, D.W.; Broders, K.; Kim, H.S.; McCormick, S.P.; Busman, M.; Aoki, T.; et al. DNA sequence-based identification of Fusarium: A work in progress. Plant Dis. 2022, 106, 1597–1609. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Rep, M.; Wang, B.; Ashton, A.; Dodds, P.; Ellis, J. Variation in potential effector genes distinguishing Australian and non-Australian isolates of the cotton wilt pathogen Fusarium oxysporum f. sp. vasinfectum. Plant Pathol. 2011, 60, 232–243. [Google Scholar] [CrossRef]
- Ortiz, C.S.; Bell, A.A.; Magill, C.W.; Liu, J. Specific PCR detection of Fusarium oxysporum f. sp. vasinfectum California race 4 based on a unique Tfo1 insertion event in the PHO gene. Plant Dis. 2017, 101, 34–44. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Hutmacher, R.; Davis, R. Characterization of California isolates of Fusarium oxysporum f. sp. vasinfectum. Plant Dis. 2005, 89, 366–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, J.; Garcia, J.; Lara, C.; Hutmacher, R.B.; Ulloa, M.; Nichols, R.L.; Ellis, M.L. Characterization of current Fusarium oxysporum f. sp. vasinfectum isolates from cotton in the San Joaquin Valley of California and Lower Valley El Paso, Texas. Plant Dis. 2021, 105, 1898–1911. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Abdelraheem, A.; Lujan, P.; Idowu, J.; Sullivan, P.; Nichols, R.; Wedegaertner, T.; Zhang, J. Detection and Characterization of Fusarium Wilt (Fusarium oxysporum f. sp. vasinfectum) Race 4 Causing Fusarium Wilt of Cotton Seedlings in New Mexico. Plant Dis. 2021, 105, 3353–3367. [Google Scholar] [CrossRef]
- Wang, B.; Brubaker, C.L.; Tate, W.; Woods, M.J.; Matheson, B.A.; Burdon, J.J. Genetic variation and population structure of Fusarium oxysporum f. sp. vasinfectum in Australia. Plant Pathol. 2006, 55, 746–755. [Google Scholar] [CrossRef]
- Smith, L. Management of Fusarium Wilt of Cotton (DAQ130C); Cotton Research and Development Corporation: Narrabri, Australia, 2007; pp. 1–25. [Google Scholar]
- Smith, L. Cotton Fusarium Wilt Management (DAQ130C); Cotton Research and Development Corporation: Narrabri, Australia, 2010; pp. 1–75. [Google Scholar]
- Bell, A.A.; Gu, A.; Olvey, J.; Wagner, T.A.; Tashpulatov, J.J.; Prom, S.; Quintana, J.; Nichols, R.L.; Liu, J. Detection and characterization of Fusarium oxysporum f. sp. vasinfectum VCG0114 (race 4) isolates of diverse geographic origins. Plant Dis. 2019, 103, 1998–2009. [Google Scholar] [CrossRef]
- Assigbetse, K.B.; Fernandez, D.; Dubois, M.P.; Geiger, J.P. Differentiation of Fusarium oxysporum f. sp. vasinfectum races on cotton by random amplified polymorphic DNA (RAPD) analysis. Phytopathology 1994, 84, 622–626. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, D.; Assigbese, K.; Dubois, M.P.; Geiger, J.P. Molecular characterization of races and vegetative compatibility groups in Fusarium oxysporum f. sp. vasinfectum. Appl. Environ. Microbiol. 1994, 60, 4039–4046. [Google Scholar] [CrossRef] [Green Version]
- Le, D.P.; Gregson, A.; Tran, T.T.; Jackson, R. Co-occurrence of defoliating and non-defoliating pathotypes of Verticillium dahliae in field-grown cotton plants in New South Wales, Australia. Plants 2020, 9, 750. [Google Scholar] [CrossRef]
- Le, D.P.; Tran, T.T.; Gregson, A.; Jackson, R. TEF1 sequence-based diversity of Fusarium species recovered from collar rot diseased cotton seedlings in New South Wales, Australia. Australas. Plant Pathol. 2020, 49, 277–284. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puhalla, J.E. Classification of strains of Fusarium oxysporum on the basis of vegetative compatibility. Canad. J. Bot. 1985, 63, 179–183. [Google Scholar] [CrossRef]
- Correll, J.; Klittich, C.; Leslie, J. Nitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests. Phytopathology 1987, 77, 1640–1646. [Google Scholar] [CrossRef]
- Puhalla, J.E.; Spieth, P.T. Heterokaryosis in Fusarium moniliforme. Exp. Mycol. 1983, 7, 328–335. [Google Scholar] [CrossRef]
- Rothrock, C.; Woodward, J.; Kemerait, R. Diseases. In Cotton (Agronomy Monograph No. 57), 2nd ed.; Fang, D., Percy, R., Eds.; American Society of Agronomy: Madison, WI, USA, 2015; pp. 465–507. [Google Scholar]
- Wang, B.; Dale, M.; Kochman, J. Studies on a pathogenicity assay for screening cotton germplasms for resistance to Fusarium oxysporum f. sp. vasinfectum in the glasshouse. Aust. J. Exp. Agric. 1999, 39, 967–974. [Google Scholar] [CrossRef]
- Liu, J.; Bell, A.A.; Wheeler, M.H.; Stipanovic, R.D.; Puckhaber, L.S. Phylogeny and pathogenicity of Fusarium oxysporum isolates from cottonseed imported from Australia into California for dairy cattle feed. Canad. J. Microbiol. 2011, 57, 874–886. [Google Scholar] [CrossRef]
- Jangir, P.; Mehra, N.; Sharma, K.; Singh, N.; Rani, M.; Kapoor, R. Secreted in xylem genes: Drivers of host adaptation in Fusarium oxysporum. Front. Plant Sci. 2021, 12, 628611. [Google Scholar] [CrossRef]
- Czislowski, E.; Zeil-Rolfe, I.; Aitken, E.A. Effector profiles of endophytic Fusarium associated with asymptomatic banana (Musa sp.) hosts. Int. J. Mol. Sci. 2021, 22, 2508. [Google Scholar] [CrossRef]
- Gawehns, F.; Houterman, P.M.; Ichou, F.A.; Michielse, C.B.; Hijdra, M.; Cornelissen, B.J.C.; Rep, M.; Takken, F.L.W. The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microbe Interact. 2014, 27, 336–348. [Google Scholar] [CrossRef]
- Rocha, L.O.; Laurence, M.H.; Ludowici, V.A.; Puno, V.I.; Lim, C.C.; Tesoriero, L.A.; Summerell, B.A.; Liew, E.C. Putative effector genes detected in Fusarium oxysporum from natural ecosystems of Australia. Plant Pathol. 2016, 65, 914–929. [Google Scholar] [CrossRef] [Green Version]
- Cooper, W.; Brodie, B. A comparison of Fusarium-wilt indices of cotton varieties with root-knot and sting nematodes as predisposing agents. Phytopathology 1963, 53, 1077–1080. [Google Scholar]
- Garber, R.; Jorgenson, E.; Smith, S.; Hyer, A. Interaction of population levels of Fusarium oxysporum f. sp. vasinfectum and Meloidogyne incognita on cotton. J. Nematol. 1979, 11, 133–137. [Google Scholar]
- Smith, L. Understanding the Ecology of Reniform Nematodes in Cotton (DAQ1803); Cotton Research and Development Corporation: Narrabri, Australia, 2019; pp. 1–32. [Google Scholar]
- Neal, D. The reniform nematode and its relationship to the incidence of Fusarium wilt of cotton at Baton-Rouge, Louisiana. Phytopathology 1954, 44, 447–450. [Google Scholar]
- Bennett, R.; Hutmacher, R.; Davis, R. Seed transmission of Fusarium oxysporum f. sp. vasinfectum race 4 in California. J. Cotton Sci. 2008, 12, 160–164. [Google Scholar]
- Dyer, D.R.; Newman, M.; Lawrence, K.S. Diversity and temporal distribution of Fusarium oxysporum f. sp. vasinfectum races and genotypes as influenced by Gossypium cultivar. Front. Fungal Biol. 2022, 3, 1022761. [Google Scholar] [CrossRef]
- Bayraktar, H.; Dolar, F.S.; Maden, S. Use of RAPD and ISSR markers in detection of genetic variation and population structure among Fusarium oxysporum f. sp. ciceris isolates on chickpea in Turkey. J. Phytopathol. 2008, 156, 146–154. [Google Scholar] [CrossRef]
- Claerbout, J.; Van Poucke, K.; Mestdagh, H.; Delaere, I.; Vandevelde, I.; Venneman, S.; Decombel, A.; Bleyaert, P.; Neukermans, J.; Viaene, N.; et al. Fusarium isolates from Belgium causing wilt in lettuce show genetic and pathogenic diversity. Plant Pathol. 2022. [Google Scholar] [CrossRef]

| Valleys 1 | Season 2017/18 | Season 2018/19 | Season 2019/20 | Season 20/21 | Season 21/22 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| FW Fields 2 | Incidence (%) 3 | FW Fields | Incidence (%) | FW Fields | Incidence (%) | FW Fields | Incidence (%) | FW Fields | Incidence (%) | |
| Emerald | 0/7 | 0 | 0/11 | 0 | na | na | na | na | 0/12 | 0 |
| Theodore | 1/11 | 1 | 1/11 | 2.5 | na | na | 1/14 | 4.7 | 5/18 | 2 (0.5–5) |
| St George | 7/10 | 6 (1.5–26) | 4/8 | 10.8 (3–30) | 4/7 | 2.3 (0.5–6) | 3/14 | 0.9 (0.5–2.3) | 2/5 | 13.8 (1–26.5) |
| Darling Downs | 1/11 | 5.5 | 10/17 | 4.7 (0.5–13.5) | 5/18 | 3.1 (0.3–9) | 8/20 | 4 (0.7–12.3) | 11/13 | 7.1 (0.5–44.5) |
| Border Rivers | na | na | 1/13 | 0.5 | 1/5 | 2.5 | 3/13 | 1.3 (0.3–3) | 7/14 | 1.5 (0.5–4) |
| Gwydir | 0/8 | 0 | 0/13 | 0 | 0/10 | 0 | 0 [34]/11 | Detected | 8 [34]/19 | 5.8 (0.3–20.5) |
| Namoi | 0/20 | 0 | 0/18 | 0 | 0/10 | 0 | 0/10 | 0 | 0/20 | 0 |
| Macquarie | 0/10 | 0 | 0/10 | 0 | 0/6 | 0 | 0/7 | 0 | 0/13 | 0 |
| Lachlan | 0/8 | 0 | 0/9 | 0 | 0/8 | 0 | 0/7 | 0 | 0/5 | 0 |
| Murrumbidgee | 0/13 | 0 | 0/14 | 0 | 0/8 | 0 | 0/11 | 0 | 0/9 | 0 |
| Valleys 1 | Season 17/18 | Season 18/19 | Season 19/20 | Season 20/21 | Season 21/22 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| FW Fields 2 | Incidence 3 (%) | FW Fields | Incidence (%) | FW Fields | Incidence (%) | FW Fields | Incidence (%) | FW Fields | Incidence (%) | |
| Emerald | 0/8 | 0 | 0/4 | 0 | na | na | na | na | 1/10 | 4.5 |
| Theodore | 3/10 | 5.7 (2–12) | 1/10 | 33 | 5/19 | 4.3 (0.5–10) | 0/7 | 0 | 2/10 | 20 (3–37) |
| St George | 7/10 | 15 (0.5–46) | 6/8 | 18 (0.5–58) | 3/6 | 1.3 (0.5–2) | 4/11 | 2.3 (0.5–3.5) | 9/16 | 27.7 (0.5–97) |
| Darling Downs | 5/10 | 5.2 (0.5–20) | 11/18 | 4.3 (0.3–21) | 6/17 | 4.9 (0.5–10) | 6/8 | 4 (0.5–14) | 10/13 | 15.9 (1.5–92) |
| Border Rivers | 7/9 | 18 (1–60) | 6/15 | 3.5 (0.5–10) | 2/5 | 1 (0.5–1.5) | 5/12 | 8.4 (0.5–20) | 10/14 | 18 (3.3–61) |
| Gwydir | 4 (3)/8 | 10.7 (0.5–50) | 7 [34]/13 | 12.9 (0.5–68) | 7 [34]/10 | 6.14 (1–39) | 9 (10)/11 | 25.5 (1.5–96) | 16 (1)/19 | 39.4 (0.5–98.5) |
| Namoi | 0/20 | 0 | 0/18 | 0 | 0/10 | 0 | 0 (1)/10 | Detected | 0 [34]/20 | Detected |
| Macquarie | 1 (1)/10 | 0.7 | 2 (3)/10 | 9.8 (3.5–14) | 0/6 | 0 | 0/7 | 0 | 0 (1)/13 | Detected |
| Lachlan | 0/8 | 0 | 0/9 | 0 | 1 [34]/8 | 0.5 | 1/7 | 1 | 0/5 | 0 |
| Murrumbidgee | 0/13 | 0 | 1 (1)/14 | 0.5 | 1/8 | 0.5 | 2 (1)/11 | 2.5 (1–4) | 0/9 | 0 |
| Valleys | Season 17/18 | Season 18/19 | Season 19/20 | Season 20/21 | Season 21/22 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| No Fusarium | No Fov | No Fusarium | No Fov | No Fusarium | No Fov | No Fusarium | No Fov | No Fusarium | No Fov | |
| Gwydir | 32 | 32 | 54 | 54 | 38 | 38 | 182 | 171 | 274 | 256 |
| Namoi | 3 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 10 | 4 |
| Macquarie | 18 | 4 | 28 | 28 | 0 | 0 | 0 | 0 | 33 | 33 |
| Lachlan | 0 | 0 | 0 | 0 | 36 | 18 | 4 | 0 | 0 | 0 |
| Murrumbidgee | 0 | 0 | 14 | 14 | 3 | 3 | 10 | 10 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, D.P.; Nguyen, C.P.T.; Kafle, D.; Scheikowski, L.; Montgomery, J.; Lambeth, E.; Thomas, A.; O’Keeffe, K.; Shakeshaft, B.; Young, A.; et al. Surveillance, Diversity and Vegetative Compatibility Groups of Fusarium oxysporum f. sp. vasinfectum Collected in Cotton Fields in Australia (2017 to 2022). Pathogens 2022, 11, 1537. https://doi.org/10.3390/pathogens11121537
Le DP, Nguyen CPT, Kafle D, Scheikowski L, Montgomery J, Lambeth E, Thomas A, O’Keeffe K, Shakeshaft B, Young A, et al. Surveillance, Diversity and Vegetative Compatibility Groups of Fusarium oxysporum f. sp. vasinfectum Collected in Cotton Fields in Australia (2017 to 2022). Pathogens. 2022; 11(12):1537. https://doi.org/10.3390/pathogens11121537
Chicago/Turabian StyleLe, Duy P., Chi P. T. Nguyen, Dinesh Kafle, Linda Scheikowski, Janelle Montgomery, Emma Lambeth, Amanda Thomas, Kieran O’Keeffe, Beth Shakeshaft, Alison Young, and et al. 2022. "Surveillance, Diversity and Vegetative Compatibility Groups of Fusarium oxysporum f. sp. vasinfectum Collected in Cotton Fields in Australia (2017 to 2022)" Pathogens 11, no. 12: 1537. https://doi.org/10.3390/pathogens11121537
APA StyleLe, D. P., Nguyen, C. P. T., Kafle, D., Scheikowski, L., Montgomery, J., Lambeth, E., Thomas, A., O’Keeffe, K., Shakeshaft, B., Young, A., Mckay, A., Twine, A., Hudson, E., Jackson, R., & Smith, L. J. (2022). Surveillance, Diversity and Vegetative Compatibility Groups of Fusarium oxysporum f. sp. vasinfectum Collected in Cotton Fields in Australia (2017 to 2022). Pathogens, 11(12), 1537. https://doi.org/10.3390/pathogens11121537







