Role of Diagnostics in Epidemiology, Management, Surveillance, and Control of Leptospirosis
Abstract
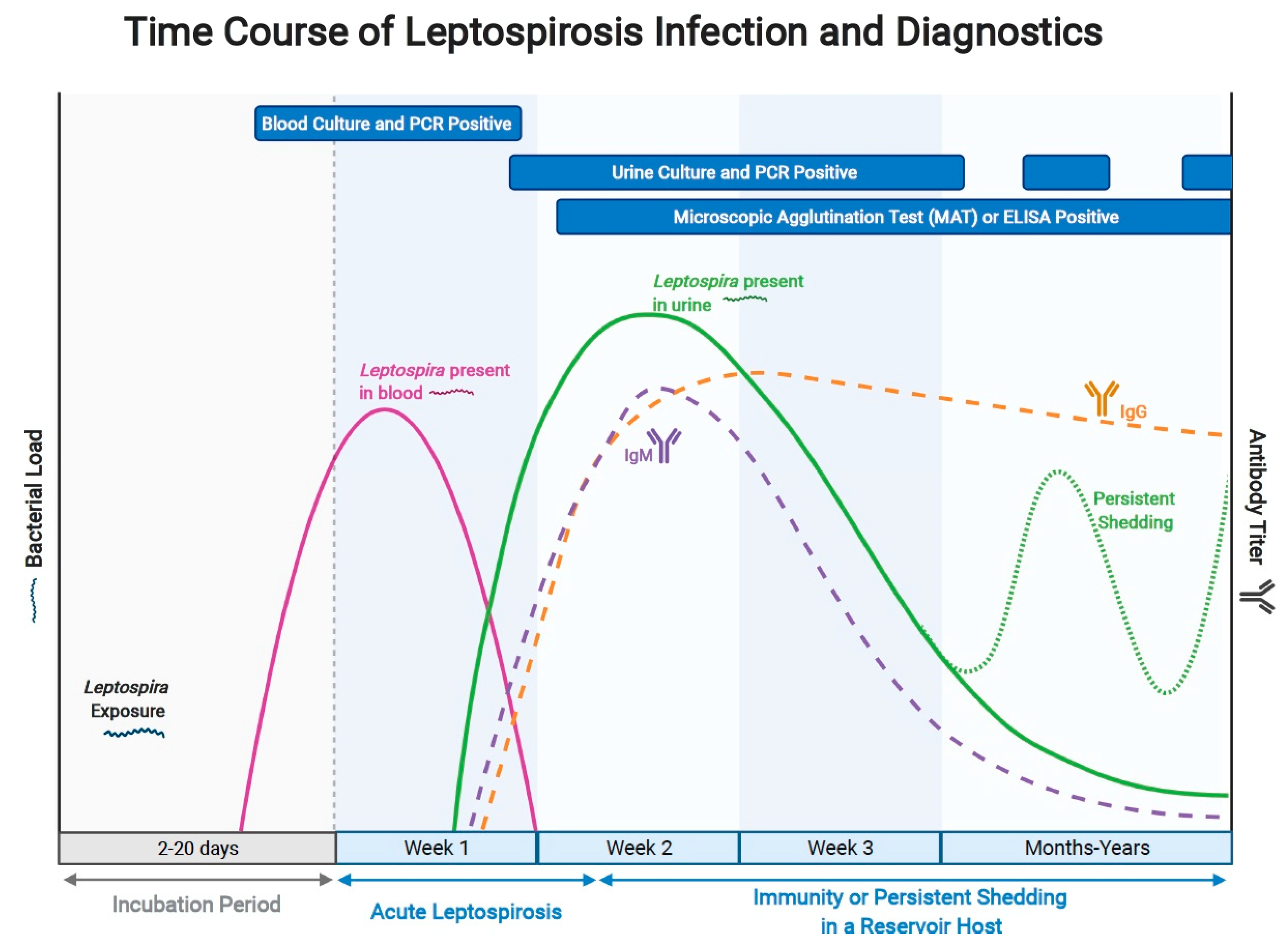
1. Introduction
2. Diagnostic Approaches—Overview
2.1. Molecular
2.2. Serology
2.3. Culture
2.4. Whole Genome Sequencing
2.5. Serotyping of Cultured Isolates
3. Diagnostics in Humans
3.1. Clinical Diagnosis
3.2. Routine Laboratory Studies and Biomarkers
3.3. Role of Diagnostics in Management of Acute Infection
3.4. Roles of Diagnostics in Human Outbreak Control
3.5. Roles of Diagnostics in Human Epidemiology and Surveillance
4. Diagnostics in Companion Animals
4.1. Cats
4.2. Epidemiology of Canine Leptospirosis
4.3. Diagnosis
5. Diagnostics in Livestock
5.1. Diagnostics and Epidemiology
5.2. Surveillance and Control
6. Future Directions
Disclaimer
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Costa, F.; Hagan, J.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef]
- Boey, K.; Shiokawa, K.; Rajeev, S. Leptospira infection in rats: A literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 2019, 13, e0007499. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.A.N.; Ribeiro, P.D.S.; da Franca, G.V.; Souza, F.N.; Ramos, E.A.G.; Figueira, C.P.; Reis, M.G.; Costa, F.; Ristow, P. Leptospira interrogans biofilm formation in Rattus norvegicus (Norway rats) natural reservoirs. PLoS Negl. Trop. Dis. 2021, 9, e0009736. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Bertelloni, F.; Albini, S.; Fratini, F. Insight into the Epidemiology of Leptospirosis: A Review of Leptospira Isolations from “Unconventional” Hosts. Animals 2021, 11, 191. [Google Scholar] [CrossRef]
- Philip, N.; Affendy, N.B.; Masri, S.N.; Yuhana, M.Y.; Than, L.T.L.; Sekawi, Z.; Neela, V.K. Combined PCR and MAT improves the early diagnosis of the biphasic illness leptospirosis. PLoS ONE 2020, 15, e0239069. [Google Scholar] [CrossRef] [PubMed]
- Nally, J.E.; Ahmed, A.A.A.; Putz, E.J.; Palmquist, D.E.; Goris, M.G.A. Comparison of Real-Time PCR, Bacteriologic Culture and Fluorescent Antibody Test for the Detection of Leptospira borgpetersenii in Urine of Naturally Infected Cattle. Vet. Sci. 2020, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Miyahara, S.; Villanueva, S.Y.; Saito, M.; Gloriani, N.G.; Yoshida, S.-I. A novel combination of selective agents for isolation of Leptospira species. Microbiol. Immunol. 2011, 55, 494–501. [Google Scholar] [CrossRef]
- Hornsby, R.L.; Alt, D.P.; Nally, J.E. Isolation and propagation of leptospires at 37 °C directly from the mammalian host. Sci. Rep. 2020, 10, 9620. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.A.; Montgomery, J.; Cassells, J.A. Dihydrostreptomycin treatment of bovine carriers of Leptospira interrogans serovar hardjo. Res. Vet. Sci. 1985, 39, 292–295. [Google Scholar] [CrossRef]
- Mancel, E.; Vetd, F.M.; Pesenti, L.; Salino, D.; Angibaud, G.; Perolat, P. Clinical aspects of ocular leptospirosis in New Caledonia (South Pacific). Aust. N. Z. J. Ophthalmol. 1999, 27, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; De Vries, S.G.; Ahmed, A.; Visser, B.J.; Nagel, I.M.; Spijker, R.; Grobusch, M.P.; Hartskeerl, R.A.; Goris, M.G.; Leeflang, M.M. Nucleic acid and antigen detection tests for leptospirosis. Cochrane Database Syst. Rev. 2019, 8, CD011871. [Google Scholar] [CrossRef]
- Matthias, M.A.; Ricaldi, J.; Cespedes, M.; Diaz, M.; Galloway, R.L.; Saito, M.; Steigerwalt, A.G.; Patra, K.P.; Ore, C.V.; Gotuzzo, E.; et al. Human Leptospirosis Caused by a New, Antigenically Unique Leptospira Associated with a Rattus Species Reservoir in the Peruvian Amazon. PLoS Negl. Trop. Dis. 2008, 2, e213. [Google Scholar] [CrossRef] [PubMed]
- Chiriboga, J.; Barragan, V.; Arroyo, G.; Sosa, A.; Birdsell, D.N.; España, K.; Mora, A.; Espín, E.; Mejía, M.E.; Al, J.C.E.; et al. High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador. Emerg. Infect. Dis. 2015, 21, 2141–2147. [Google Scholar] [CrossRef]
- Balamurugan, V.; Gangadhar, N.L.; Mohandoss, N.; Thirumalesh, S.R.A.; Dhar, M.; Shome, R.; Krishnamoorthy, P.; Prabhudas, K.; Rahman, H. Characterization of Leptospira isolates from animals and humans: Phylogenetic analysis identifies the prevalence of intermediate species in India. SpringerPlus 2013, 2, 362. [Google Scholar] [CrossRef] [PubMed]
- Puche, R.; Ferrés, I.; Caraballo, L.; Rangel, Y.; Picardeau, M.; Takiff, H.; Iraola, G. Leptospira venezuelensis sp. nov., a new member of the intermediate group isolated from rodents, cattle and humans. Int. J. Syst. Evol. Microbiol. 2018, 68, 513–517. [Google Scholar] [CrossRef]
- Levett, P.N.; Morey, R.E.; Galloway, R.L.; Steigerwalt, A.G. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int. J. Syst. Evol. Microbiol. 2006, 56, 671–673. [Google Scholar] [CrossRef]
- International Proficiency Testing Scheme for the Leptospirosis MAT. Available online: http://leptosociety.org/proficiency_testing/ (accessed on 4 November 2021).
- Smythe, L.D.; Tiengrim, S.; Peacock, S.J.; Wuthiekanun, V.; Suputtamongkol, Y.; Day, N.P.; Apiwattanaporn, A.; Chierakul, W.; Slack, A.T.; Chueasuwanchai, S.; et al. The Microscopic Agglutination Test (MAT) Is an Unreliable Predictor of Infecting Leptospira Serovar in Thailand. Am. J. Trop. Med. Hyg. 2009, 81, 695–697. [Google Scholar] [CrossRef]
- Murray, C.K.; Gray, M.R.; Mende, K.; Parker, T.M.; Samir, A.; Rahman, B.A.; Habashy, E.E.; Hospenthal, D.R.; Pimentel, G. Use of patient-specific Leptospira isolates in the diagnosis of leptospirosis employing microscopic agglutination testing (MAT). Trans. R. Soc. Trop. Med. Hyg. 2011, 105, 209–213. [Google Scholar] [CrossRef]
- Levett, P.N. Usefulness of Serologic Analysis as a Predictor of the Infecting Serovar in Patients with Severe Leptospirosis. Clin. Infect. Dis. 2003, 36, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M.; Bertherat, E.; Jancloes, M.; Skouloudis, A.N.; Durski, K.; Hartskeerl, R.A. Rapid tests for diagnosis of leptospirosis: Current tools and emerging technologies. Diagn. Microbiol. Infect. Dis. 2014, 78, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Niloofa, R.; Karunanayake, L.; de Silva, H.J.; Premawansa, S.; Rajapakse, S.; Handunnetti, S. Development of in-house ELISAs as an alternative method for the serodiagnosis of leptospirosis. Int. J. Infect. Dis. 2021, 105, 135–140. [Google Scholar] [CrossRef]
- Zin, N.M.; Othman, S.N.; Abd Rahman, F.R.; Abdul Rachman, A.R. Evaluation of IgM LAT and IgM ELISA as compared to microscopic agglutination test (MAT) for early diagnosis of Leptospira sp. Trop. Biomed. 2019, 36, 1071–1080. [Google Scholar] [PubMed]
- Terpstra, W.J. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control; World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Picardeau, M. Diagnosis and epidemiology of leptospirosis. Méd. Mal. Infect. 2013, 43, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bajani, M.D.; Ashford, D.A.; Bragg, S.L.; Woods, C.W.; Aye, T.; Spiegel, R.A.; Plikaytis, B.D.; Perkins, B.A.; Phelan, M.; Levett, P.N.; et al. Evaluation of four commercially available rapid serologic tests for diagnosis of leptospirosis. J. Clin. Microbiol. 2003, 41, 803–809. [Google Scholar] [CrossRef]
- Cumberland, P.; Everard, C.O.; Levett, P.N. Assessment of the efficacy of an IgM-elisa and microscopic agglutination test (MAT) in the diagnosis of acute leptospirosis. Am. J. Trop. Med. Hyg. 1999, 61, 731–734. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef]
- Signorini, M.L.; Lottersberger, J.; Tarabla, H.D.; Vanasco, N.B. Enzyme-linked immunosorbent assay to diagnose human leptospirosis: A meta-analysis of the published literature. Epidemiol. Infect. 2012, 141, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Fiecek, B.; Chmielewski, T.; Sadkowska-Todys, M.; Czerwinski, M.; Zalewska, G.; Roguska, U.; Tylewska-Wierzbanowska, S. An outbreak of leptospirosis imported from Germany to Poland. Adv. Clin. Exp. Med. 2017, 26, 415–419. [Google Scholar] [CrossRef]
- Cassadou, S.; Rosine, J.; Flamand, C.; Escher, M.; Ledrans, M.; Bourhy, P.; Picardeau, M.; Quénel, P. Underestimation of Leptospirosis Incidence in the French West Indies. PLoS Negl. Trop. Dis. 2016, 10, e0004668. [Google Scholar] [CrossRef]
- Pakoa, J.G.; Soupé-Gilbert, M.-E.; Girault, D.; Takau, D.; Gaviga, J.; Gourinat, A.-C.; Tarantola, A.; Goarant, C. High incidence of leptospirosis in an observational study of hospital outpatients in Vanuatu highlights the need for improved awareness and diagnostic capacities. PLoS Negl. Trop. Dis. 2018, 12, e0006564. [Google Scholar] [CrossRef] [PubMed]
- Limmathurotsakul, D.; Turner, E.L.; Wuthiekanun, V.; Thaipadungpanit, J.; Suputtamongkol, Y.; Chierakul, W.; Smythe, L.D.; Day, N.P.J.; Cooper, B.; Peacock, S.J. Fool’s Gold: Why Imperfect Reference Tests Are Undermining the Evaluation of Novel Diagnostics: A Reevaluation of 5 Diagnostic Tests for Leptospirosis. Clin. Infect. Dis. 2012, 55, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Schofield, M.R.; Maze, M.J.; Crump, J.A.; Rubach, M.P.; Galloway, R.; Sharples, K.J. On the robustness of latent class models for diagnostic testing with no gold standard. Stat. Med. 2021, 40, 4751–4763. [Google Scholar] [CrossRef] [PubMed]
- Lessa-Aquino, C.; Rodrigues, C.B.; Pablo, J.; Sasaki, R.; Jasinskas, A.; Liang, L.; Wunder, E.A.; Ribeiro, G.S.; Vigil, A.; Galler, R.; et al. Identification of Seroreactive Proteins of Leptospira interrogans Serovar Copenhageni Using a High-Density Protein Microarray Approach. PLoS Negl. Trop. Dis. 2013, 7, e2499. [Google Scholar] [CrossRef] [PubMed]
- Nabity, S.A.; Ribeiro, G.S.; Aquino, C.L.; Takahashi, D.; Damião, A.O.; Gonçalves, A.H.O.; Miranda-Filho, D.B.; Greenwald, R.; Esfandiari, J.; Lyashchenko, K.P.; et al. Accuracy of a Dual Path Platform (DPP) Assay for the Rapid Point-of-Care Diagnosis of Human Leptospirosis. PLoS Negl. Trop. Dis. 2012, 6, e1878. [Google Scholar] [CrossRef]
- McBride, A.J.; Cerqueira, G.M.; Suchard, M.A.; Moreira, A.N.; Zuerner, R.L.; Reis, M.G.; Haake, D.A.; Ko, A.I.; Dellagostin, O.A. Genetic diversity of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect. Genet. Evol. 2009, 9, 196–205. [Google Scholar] [CrossRef]
- Goris, M.G.A.; Leeflang, M.M.G.; Boer, K.R.; Goeijenbier, M.; van Gorp, E.C.; Wagenaar, J.F.P.; Hartskeerl, R. Establishment of valid laboratory case definition for human leptospirosis. J. Bacteriol. Parasitol. 2012, 3, 132. [Google Scholar]
- Dawson, P.; Marfel, M.; Galloway, R.; Tareg, A.; Paz-Bailey, G.; Muñoz-Jordán, J.L.; Sharp, T.M.; Adams, L.E.; Bower, W.A. Notes from the Field: Interpretation of Rapid Diagnostic Tests for Leptospirosis During a Dengue Outbreak—Yap State, Federated States of Micronesia, 2019. MMWR Morb. Mortal. Wkly. Rep. 2020, 69, 1832–1833. [Google Scholar] [CrossRef]
- Miller, D.A.; Wilson, M.A.; Beran, G.W. Survey to estimate prevalence of Leptospira interrogans infection in mature cattle in the United States. Am. J. Vet. Res. 1991, 52, 1761–1765. [Google Scholar] [PubMed]
- Nally, J.E.; Hornsby, R.L.; Alt, D.; Bayles, D.; Wilson-Welder, J.; Palmquist, D.E.; Bauer, N.E. Isolation and characterization of pathogenic leptospires associated with cattle. Vet. Microbiol. 2018, 218, 25–30. [Google Scholar] [CrossRef]
- Nally, J.E.; Mullen, W.; Callanan, J.J.; Mischak, H.; Albalat, A. Detection of urinary biomarkers in reservoir hosts of leptospirosis by capillary electrophoresis-mass spectrometry. Proteom.-Clin. Appl. 2015, 9, 543–551. [Google Scholar] [CrossRef]
- Subharat, S.; Wilson, P.; Heuer, C.; Collins-Emerson, J.; Smythe, L.; Dohnt, M.; Craig, S.; Burns, M. Serosurvey of leptospirosis and investigation of a possible novel serovar Arborea in farmed deer in New Zealand. N. Z. Vet. J. 2011, 59, 139–142. [Google Scholar] [CrossRef]
- Piredda, I.; Ponti, M.N.; Piras, A.; Palmas, B.; Pintore, P.; Pedditzi, A.; Chisu, V. New Insights on Leptospira Infections in a Canine Population from North Sardinia, Italy: A Sero-Epidemiological Study. Biology 2021, 10, 507. [Google Scholar] [CrossRef]
- Narkkul, U.; Thaipadungpanit, J.; Srilohasin, P.; Singkhaimuk, P.; Thongdee, M.; Chaiwattanarungruengpaisan, S.; Krairojananan, P.; Pan-Ngum, W. Optimization of Culture Protocols to Isolate Leptospira spp. from Environmental Water, Field Investigation, and Identification of Factors Associated with the Presence of Leptospira spp. in the Environment. Trop. Med. Infect. Dis. 2020, 5, 94. [Google Scholar] [CrossRef]
- Thaipadungpanit, J.; Wuthiekanun, V.; Chierakul, W.; Smythe, L.D.; Petkanchanapong, W.; Limpaiboon, R.; Apiwatanaporn, A.; Slack, A.T.; Suputtamongkol, Y.; White, N.J.; et al. A Dominant Clone of Leptospira interrogans Associated with an Outbreak of Human Leptospirosis in Thailand. PLoS Negl. Trop. Dis. 2007, 1, e56. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.R.; Buchholz, A.E.; Hinson, K.; Park, S.Y.; Effler, P.V. Leptospirosis in Hawaii, USA, 1999–2008. Emerg. Infect. Dis. 2011, 17, 221–226. [Google Scholar] [CrossRef]
- Cranford, H.M.; Browne, A.S.; LeCount, K.; Anderson, T.; Hamond, C.; Schlater, L.; Stuber, T.; Burke-France, V.J.; Taylor, M.; Harrison, C.J.; et al. Mongooses (Urva auropunctata) as reservoir hosts of Leptospira species in the United States Virgin Islands, 2019–2020. PLoS Negl. Trop. Dis. 2021, 15, e0009859. [Google Scholar] [CrossRef]
- Levett, P.N. 3.14 Leptospira Culture. In Clinical Microbiology Procedures Handbook, 4th ed.; Leber, A.L., Ed.; ASM Press: Washington, DC, USA, 2016. [Google Scholar] [CrossRef]
- Haake, D.A.; Dundoo, M.; Cader, R.; Kubak, B.M.; Hartskeerl, R.A.; Sejvar, J.J.; Ashford, D.A. Leptospirosis, water sports, and chemoprophylaxis. Clin. Infect. Dis. 2002, 34, e40–e43. [Google Scholar] [CrossRef]
- Zarantonelli, L.; Suanes, A.; Meny, P.; Buroni, F.; Nieves, C.; Salaberry, X.; Briano, C.; Ashfield, N.; Silveira, C.D.S.; Dutra, F.; et al. Isolation of pathogenic Leptospira strains from naturally infected cattle in Uruguay reveals high serovar diversity, and uncovers a relevant risk for human leptospirosis. PLoS Negl. Trop. Dis. 2018, 12, e0006694. [Google Scholar] [CrossRef]
- Miller, D.A.; Wilson, M.A.; Beran, G.W. The effect of storage time on isolation of Leptospira interrogans from bovine kidneys. J. Vet. Diagn. Investig. 1990, 2, 63–65. [Google Scholar] [CrossRef]
- Guedes, I.B.; Rocha, K.D.S.; Negrão, M.P.; de Souza, G.O.; Castro, J.F.D.P.; Cavalini, M.B.; Filho, A.F.D.S.; Neto, M.S.D.; Aizawa, J.; de Moraes, C.C.G.; et al. Leptospira transport medium (LTM): A practical tool for leptospires isolation. J. Microbiol. Methods 2020, 175, 105995. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.F.; Blackmore, D.K. Effect of chilling and freezing on survival of Leptospira interrogans serovar pomona in naturally infected pig kidneys. N. Z. Vet. J. 1979, 27, 121–123. [Google Scholar] [CrossRef]
- Schürch, A.; Arredondo-Alonso, S.; Willems, R.; Goering, R. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene–based approaches. Clin. Microbiol. Infect. 2018, 24, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Guglielmini, J.; Bourhy, P.; Schiettekatte, O.; Zinini, F.; Brisse, S.; Picardeau, M. Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PLoS Negl. Trop. Dis. 2019, 13, e0007374. [Google Scholar] [CrossRef]
- Faine, S.; Adler, B.; Bolin, C.; Perolat, P. Leptospira and Leptospirosis, 2nd ed.; MediSci: Melbourne, Australia, 1999. [Google Scholar]
- Brenner, D.J.; Kaufmann, A.F.; Sulzer, K.R.; Steigerwalt, A.G.; Rogers, F.C.; Weyant, R.S. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 1999, 49, 839–858. [Google Scholar] [CrossRef]
- Kmety, E.; Dikken, H. Classification of the Species Leptospira Interrogans and History of Its Serovars; University Press Groningen: Groningen, The Netherlands, 1993. [Google Scholar]
- Miraglia, F.; Matsuo, M.; Morais, Z.M.; Dellagostin, O.A.; Seixas, F.K.; Freitas, J.C.; Hartskeerl, R.; Moreno, L.; Costa, B.L.; Souza, G.O.; et al. Molecular characterization, serotyping, and antibiotic susceptibility profile of Leptospira interrogans serovar Copenhageni isolates from Brazil. Diagn. Microbiol. Infect. Dis. 2013, 77, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Mgode, G.; Machang’U, R.S.; Mhamphi, G.G.; Katakweba, A.; Mulungu, L.S.; Durnez, L.; Leirs, H.; Hartskeerl, R.A.; Belmain, S. Leptospira Serovars for Diagnosis of Leptospirosis in Humans and Animals in Africa: Common Leptospira Isolates and Reservoir Hosts. PLoS Negl. Trop. Dis. 2015, 9, e0004251. [Google Scholar] [CrossRef] [PubMed]
- Typing with Monoclonal Antibodies. Leptospirosis Reference Centre, Amsterdam University Medical Centers. Available online: https://leptospira.amsterdamumc.org/leptospirosis-reference-centre/typing-with-monoclonal-antibodies/ (accessed on 1 January 2022).
- Leptospirosis Reference Centre, Amsterdam University Medical Centers. Available online: https://leptospira.amsterdamumc.org/leptospirosis-reference-centre/ (accessed on 1 January 2022).
- Haake, D.A.; Levett, P.N. Leptospirosis in Humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar] [CrossRef] [PubMed]
- Monahan, A.; Miller, I.; Nally, J. Leptospirosis: Risks during recreational activities. J. Appl. Microbiol. 2009, 107, 707–716. [Google Scholar] [CrossRef]
- Kumar, S.S. Indian guidelines for the diagnosis and management of human leptospirosis. In India: Medicine Update; Muruganathan, A., Ed.; The Association of Physicians of India: Mumbai, India, 2013; pp. 23–29. [Google Scholar]
- WHO. Informal Expert Consultation on Surveillance, Diagnosis and Risk Reduction of Leptospirosis. Chennai, India, 17–18 September 2009; SEA-CD-217; World Health Organization—Regional Office for South-East Asia: New Delhi, India. Available online: https://tinyurl.com/4kdnnsz7 (accessed on 23 February 2022).
- Chierakul, W.; Tientadakul, P.; Suputtamongkol, Y.; Wuthiekanun, V.; Phimda, K.; Limpaiboon, R.; Opartkiattikul, N.; White, N.J.; Peacock, S.J.; Day, N.P. Activation of the Coagulation Cascade in Patients with Leptospirosis. Clin. Infect. Dis. 2008, 46, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Wagenaar, J.F.P.; Goris, M.G.A.; Partiningrum, D.L.; Isbandrio, B.; Hartskeerl, R.A.; Brandjes, D.P.M.; Meijers, J.C.M.; Gasem, M.H.; van Gorp, E.C.M. Coagulation disorders in patients with severe leptospirosis are associated with severe bleeding and mortality. Trop. Med. Int. Health 2010, 15, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Lubell, Y.; Blacksell, S.D.; Dunachie, S.; Tanganuchitcharnchai, A.; Althaus, T.; Watthanaworawit, W.; Paris, D.H.; Mayxay, M.; Peto, T.J.; Dondorp, A.M.; et al. Performance of C-reactive protein and procalcitonin to distinguish viral from bacterial and malarial causes of fever in Southeast Asia. BMC Infect. Dis. 2015, 15, 511 . [Google Scholar] [CrossRef]
- Wangrangsimakul, T.; Althaus, T.; Mukaka, M.; Kantipong, P.; Wuthiekanun, V.; Chierakul, W.; Blacksell, S.D.; Day, N.P.; Laongnualpanich, A.; Paris, D.H. Causes of acute undifferentiated fever and the utility of biomarkers in Chiangrai, northern Thailand. PLoS Negl. Trop. Dis. 2018, 12, e0006477. [Google Scholar] [CrossRef] [PubMed]
- Tillekeratne, L.G.; Suchindran, S.; Ko, E.R.; Petzold, E.A.; Bodinayake, C.K.; Nagahawatte, A.; Devasiri, V.; Kurukulasooriya, R.; Nicholson, B.P.; McClain, M.T.; et al. Previously Derived Host Gene Expression Classifiers Identify Bacterial and Viral Etiologies of Acute Febrile Respiratory Illness in a South Asian Population. Open Forum Infect. Dis. 2020, 7, ofaa194. [Google Scholar] [CrossRef] [PubMed]
- Le Turnier, P.; Bonifay, T.; Mosnier, E.; Schaub, R.; Jolivet, A.; Demar, M.P.; Bourhy, P.; Nacher, M.; Djossou, F.; Epelboin, L. Usefulness of C-Reactive Protein in Differentiating Acute Leptospirosis and Dengue Fever in French Guiana. Open Forum Infect. Dis. 2019, 6, ofz323. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, J.; Faucher, J.-F.; Toubin, M.; Hoen, B.; Estavoyer, J.-M. Serum C-reactive protein (CRP) and procalcitonin (PCT) levels and kinetics in patients with leptospirosis. Eur. J. Clin. Microbiol. 2010, 30, 299–302. [Google Scholar] [CrossRef]
- Lindow, J.C.; Wunder, E.A., Jr.; Popper, S.J.; Min, J.-N.; Mannam, P.; Srivastava, A.; Yao, Y.; Hacker, K.; Raddassi, K.; Lee, P.J.; et al. Cathelicidin Insufficiency in Patients with Fatal Leptospirosis. PLoS Pathog. 2016, 12, e1005943. [Google Scholar] [CrossRef]
- Agampodi, S.; Matthias, M.A.; Moreno, A.C.; Vinetz, J.M. Utility of Quantitative Polymerase Chain Reaction in Leptospirosis Diagnosis: Association of Level of Leptospiremia and Clinical Manifestations in Sri Lanka. Clin. Infect. Dis. 2012, 54, 1249–1255. [Google Scholar] [CrossRef]
- BioFire Global Fever Panel—Instructions for Use. Available online: https://www.biofiredefense.com/wp-content/uploads/2021/08/BioFire-Global-Fever-Panel-IFU-DFA2-PRT-0026–02.pdf (accessed on 1 January 2022).
- Goris, M.G.A.; Leeflang, M.; Loden, M.; Wagenaar, J.F.P.; Klatser, P.; Hartskeerl, R.A.; Boer, K.R. Prospective Evaluation of Three Rapid Diagnostic Tests for Diagnosis of Human Leptospirosis. PLoS Negl. Trop. Dis. 2013, 7, e2290. [Google Scholar] [CrossRef]
- Maze, M.J.; Sharples, K.J.; Allan, K.J.; Rubach, M.; Crump, J.A. Diagnostic accuracy of leptospirosis whole-cell lateral flow assays: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2018, 25, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Nabity, S.A.; Hagan, J.; Araújo, G.; Damião, A.O.; Cruz, J.S.; Nery, N.; Wunder, E.A., Jr.; Reis, M.G.; Ko, A.; Ribeiro, G.S. Prospective evaluation of accuracy and clinical utility of the Dual Path Platform (DPP) assay for the point-of-care diagnosis of leptospirosis in hospitalized patients. PLoS Negl. Trop. Dis. 2018, 12, e0006285. [Google Scholar] [CrossRef]
- Alia, S.N.; Joseph, N.; Philip, N.; Azhari, N.N.; Garba, B.; Masri, S.N.; Sekawi, Z.; Neela, V.K. Diagnostic accuracy of rapid diagnostic tests for the early detection of leptospirosis. J. Infect. Public Health 2018, 12, 263–269. [Google Scholar] [CrossRef]
- Fornazari, F.; Richini-Pereira, V.B.; Joaquim, S.F.; Nachtigall, P.G.; Langoni, H. Leptospirosis diagnosis among patients suspected of dengue fever in Brazil. J. Venom. Anim. Toxins Incl. Trop. Dis. 2021, 27, e20200118. [Google Scholar] [CrossRef] [PubMed]
- Ittyachen, A.M. COVID-19 and leptospirosis: Cytokine storm and the use of steroids. Trop. Dr. 2020, 51, 128–130. [Google Scholar] [CrossRef]
- Zaki, S.R.; Shieh, W.J. Leptospirosis associated with outbreak of acute febrile illness and pulmonary haemorrhage, Nicaragua, 1995. The Epidemic Working Group at Ministry of Health in Nicaragua. Lancet 1996, 347, 535–536. [Google Scholar] [CrossRef]
- Trevejo, R.T.; Rigau-Perez, J.G.; Ashford, D.A.; McClure, E.M.; Jarquin-Gonzalez, C.; Amador, J.J.; de los Reyes, J.O.; Gonzalez, A.; Zaki, S.R.; Shieh, W.J.; et al. Epidemic leptospirosis associated with pulmonary hemorrhage-Nicaragua, 1995. J. Infect. Dis. 1998, 178, 1457–1463. [Google Scholar] [CrossRef]
- Romero, E.C.; Billerbeck, A.E.; Lando, V.S.; Camargo, E.D.; Souza, C.C.; Yasuda, P.H. Detection of Leptospira DNA in patients with aseptic meningitis by PCR. J. Clin. Microbiol. 1998, 36, 1453–1455. [Google Scholar] [CrossRef]
- Wilson, M.; Naccache, S.N.; Samayoa, E.; Biagtan, M.; Bashir, H.; Yu, G.; Salamat, S.M.; Somasekar, S.; Federman, S.; Miller, S.; et al. Actionable Diagnosis of Neuroleptospirosis by Next-Generation Sequencing. N. Engl. J. Med. 2014, 370, 2408–2417. [Google Scholar] [CrossRef]
- Sergio, A.D.M.; Figueroa, E.G.; Saadia, V.G.M.; Elizabeth, S.H.; Beatriz, R.S.; Víctor, M.A.A.; Espinosa, J.N. Leptospirosis Prevalence in Patients with Initial Diagnosis of Dengue. J. Trop. Med. 2012, 2012, 519701. [Google Scholar] [CrossRef][Green Version]
- Bruce, M.G.; Sanders, E.; Leake, J.; Zaidel, O.; Bragg, S.; Aye, T.; Shutt, K.; Deseda, C.; Rigau-Perez, J.; Tappero, J.; et al. Leptospirosis among patients presenting with dengue-like illness in Puerto Rico. Acta Trop. 2005, 96, 36–46. [Google Scholar] [CrossRef]
- del Valle-Mendoza, J.; Palomares-Reyes, C.; Carrillo-Ng, H.; Tarazona-Castro, Y.; Kym, S.; Aguilar-Luis, M.A.; del Valle, L.J.; Aquino-Ortega, R.; Martins-Luna, J.; Peña-Tuesta, I.; et al. Leptospirosis in febrile patients with suspected diagnosis of dengue fever. BMC Res. Notes 2021, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Larocque, R.C.; Breiman, R.F.; Ari, M.D.; Morey, R.E.; Janan, F.A.; Hayes, J.M.; Hossain, M.A.; Brooks, W.A.; Levett, P.N. Leptospirosis during Dengue Outbreak, Bangladesh. Emerg. Infect. Dis. 2005, 11, 766–769. [Google Scholar] [CrossRef]
- Nhan, T.-X.; Bonnieux, E.; Rovery, C.; De Pina, J.-J.; Musso, D. Fatal leptospirosis and chikungunya co-infection: Do not forget leptospirosis during chikungunya outbreaks. IDCases 2016, 5, 12–14. [Google Scholar] [CrossRef]
- Md-Lasim, A.; Mohd-Taib, F.S.; Abdul-Halim, M.; Mohd-Ngesom, A.M.; Nathan, S.; Md-Nor, S. Leptospirosis and Coinfection: Should We Be Concerned? Int. J. Environ. Res. Public Health 2021, 18, 9411. [Google Scholar] [CrossRef]
- Niriella, M.A.; Ediriweera, D.S.; De Silva, A.P.; Premarathna, B.H.R.; Jayasinghe, S.; de Silva, H.J. Dengue and leptospirosis infection during the coronavirus 2019 outbreak in Sri Lanka. Trans. R. Soc. Trop. Med. Hyg. 2021, 115, 944–946. [Google Scholar] [CrossRef]
- Krol, P.; Coolen-Allou, N.; Teysseyre, L.; Traversier, N.; Beasley, F.; Nativel, M.; Allou, N.; Allyn, J. Differential diagnoses of severe COVID-19 in tropical areas: The experience of Reunion Island. Trop. Med. Int. Health 2020, 26, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Bao, C. Disparity in clinical characteristics between 2019 novel coronavirus pneumonia and leptospirosis. Open Med. 2021, 16, 494–497. [Google Scholar] [CrossRef]
- Bryson, D.G.; Ellis, W.A. Leptospirosis in a British domestic cat. J. Small Anim. Pract. 1976, 17, 459–465. [Google Scholar] [CrossRef]
- Rees, H.G. Leptospirosis in a cat. N. Z. Vet. J. 1964, 12, 64. [Google Scholar] [CrossRef]
- Agunloye, C.A.; Nash, A.S. Investigation of possible leptospiral infection in cats in Scotland. J. Small Anim. Pract. 1996, 37, 126–129. [Google Scholar] [CrossRef]
- Dickeson, D.; Love, D.N. A serological survey of dogs, cats and horses in south-eastern Australia for leptospiral antibodies. Aust. Vet. J. 1993, 70, 389–390. [Google Scholar] [CrossRef] [PubMed]
- Markovich, J.; Ross, L.; McCobb, E. The Prevalence of Leptospiral Antibodies in Free Roaming Cats in Worcester County, Massachusetts. J. Vet. Intern. Med. 2012, 26, 688–689. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, M.E.; Bourtzi-Hatzopoulou, E.; Koutinas, A.F.; Petridou, E.; Saridomichelakis, M.N.; Leontides, L.; Siochu, A. Leptospiral seroepidemiology in a feline hospital population in Greece. Vet. Rec. 2005, 156, 615–616. [Google Scholar] [CrossRef]
- Rodriguez, J.; Blais, M.-C.; Lapointe, C.; Arsenault, J.; Carioto, L.; Harel, J. Serologic and Urinary PCR Survey of Leptospirosis in Healthy Cats and in Cats with Kidney Disease. J. Vet. Intern. Med. 2014, 28, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Weis, S.; Rettinger, A.; Bergmann, M.; Llewellyn, J.R.; Pantchev, N.; Straubinger, R.; Hartmann, K. Detection of Leptospira DNA in urine and presence of specific antibodies in outdoor cats in Germany. J. Feline Med. Surg. 2016, 19, 470–476. [Google Scholar] [CrossRef]
- Obrenovic, S.; Radojicic, S.; Stevic, N.; Bogunovic, D.; Vakanjac, S.; Valcic, M. Seroprevalence of cat leptospirosis in Belgrade (Serbia). Acta Vet.-Beogr. 2014, 64, 510–518. [Google Scholar]
- Dybing, N.A.; Jacobson, C.; Irwin, P.; Algar, D.; Adams, P.J. Leptospira Species in Feral Cats and Black Rats from Western Australia and Christmas Island. Vector-Borne Zoonotic Dis. 2017, 17, 319–324. [Google Scholar] [CrossRef]
- Chan, K.-W.; Hsu, Y.-H.; Hu, W.-L.; Pan, M.-J.; Lai, J.; Huang, K.-C.; Chou, S.-J. Serological and PCR Detection of Feline Leptospira in Southern Taiwan. Vector-Borne Zoonotic Dis. 2014, 14, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Gomard, Y.; Lagadec, E.; Humeau, L.; Pinet, P.; Bureau, S.; Da Silva, D.; Turpin, M.; Mattoir, Y.S.; Georger, S.; Mavingui, P.; et al. Feral cats do not play a major role in leptospirosis epidemiology on Reunion Island. Epidemiol. Infect. 2019, 147, e97. [Google Scholar] [CrossRef] [PubMed]
- Grimm, K.; Rivera, N.A.; Fredebaugh-Siller, S.; Weng, H.Y.; Warner, R.E.; Maddox, C.W.; Mateus-Pinilla, N.E. Evidence of Leptospira Serovars in Wildlife and Leptospiral DNA in Water Sources in a Natural Area in East-Central Illinois, USA. J. Wildl. Dis. 2020, 56, 316–327. [Google Scholar] [CrossRef]
- Arbour, J.; Blais, M.-C.; Carioto, L.; Sylvestre, D. Clinical Leptospirosis in Three Cats (2001–2009). J. Am. Anim. Hosp. Assoc. 2012, 48, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Beaudu-Lange, C.; Lange, E. Unusual clinical presentation of leptospirosis in a cat. Rev. Vétérinaire Clin. 2014, 49, 115–122. [Google Scholar] [CrossRef]
- Ward, M.P.; Glickman, L.T.; Guptill, L.E. Prevalence of and risk factors for leptospirosis among dogs in the United States and Canada: 677 cases (1970–1998). J. Am. Vet. Med. Assoc. 2002, 220, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Greene, C.E.; Sykes, J.E.; Brown, C.A.; Hartmann, K. Leptospirosis. In Infectious Disease of the Dog and Cat, 3rd ed.; Greene, C.E., Ed.; Saunders/Elsevier: Philadelphia, PA, USA, 2006; pp. 402–417. [Google Scholar]
- Brown, C.A.; Roberts, A.W.; Miller, M.A.; Davis, D.A.; Brown, S.A.; Bolin, C.A.; Jarecki-Black, J.; Greene, C.E.; Miller-Liebl, D. Leptospira interrogans serovar grippotyphosa infection in dogs. J. Am. Vet. Med. Assoc. 1996, 209, 1265–1267. [Google Scholar] [PubMed]
- Nielsen, J.N.; Cochran, G.K.; Cassells, J.A.; Hanson, L.E. Leptospira interrogans serovar bratislava infection in two dogs. J. Am. Vet. Med. Assoc. 1991, 199, 351–352. [Google Scholar] [PubMed]
- Stokes, J.E.; Kaneene, J.B.; Schall, W.D.; Kruger, J.M.; Miller, R.; Kaiser, L.; Bolin, C.A. Prevalence of serum antibodies against six Leptospira serovars in healthy dogs. J. Am. Vet. Med. Assoc. 2007, 230, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Gautam, R.; Wu, C.-C.; Guptill, L.F.; Potter, A.; Moore, G.E. Detection of antibodies against Leptospira serovars via microscopic agglutination tests in dogs in the United States, 2000–2007. J. Am. Vet. Med. Assoc. 2010, 237, 293–298. [Google Scholar] [CrossRef]
- Moore, G.E.; Guptill, L.F.; Glickman, N.W.; Caldanaro, R.J.; Aucoin, D.; Glickman, L.T. Canine Leptospirosis, United States, 2002–2004. Emerg. Infect. Dis. 2006, 12, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, J.R.; Krupka-Dyachenko, I.; Rettinger, A.L.; Dyachenko, V.; Stamm, I.; Kopp, P.A.; Straubinger, R.K.; Hartmann, K. Urinary shedding of leptospires and presence of Leptospira antibodies in healthy dogs from Upper Bavaria. Berl. Munch. Tierarztl. Wochenschr. 2016, 129, 251–257. [Google Scholar] [PubMed]
- White, A.M.; Zambrana-Torrelio, C.; Allen, T.; Rostal, M.K.; Wright, A.K.; Ball, E.C.; Daszak, P.; Karesh, W. Hotspots of canine leptospirosis in the United States of America. Vet. J. 2017, 222, 29–35. [Google Scholar] [CrossRef]
- Iverson, S.A.; Levy, C.; Yaglom, H.D.; Venkat, H.L.; Artus, A.; Galloway, R.; Guagliardo, S.A.J.; Reynolds, L.; Kretschmer, M.J.; Jenni, M.E.L.; et al. Clinical, diagnostic, and epidemiological features of a community-wide outbreak of canine leptospirosis in a low-prevalence region (Maricopa County, Arizona). J. Am. Vet. Med. Assoc. 2021, 258, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Jenni, M.L.; Woodward, P.; Yaglom, H.; Levy, C.; Iverson, S.A.; Kretschmer, M.; Jarrett, N.; Dooley, E.; Narang, J.; Venkat, H. Knowledge, attitudes, and practices among veterinarians during an outbreak of canine leptospirosis—Maricopa County, Arizona, 2017. Prev. Vet. Med. 2019, 172, 104779. [Google Scholar] [CrossRef] [PubMed]
- Rojas, P.-D.; Monahan, A.M.; Schuller, S.; Miller, I.S.; Markey, B.K.; Nally, J.E. Detection and quantification of leptospires in urine of dogs: A maintenance host for the zoonotic disease leptospirosis. Eur. J. Clin. Microbiol. 2010, 29, 1305–1309. [Google Scholar] [CrossRef]
- Altheimer, K.; Jongwattanapisan, P.; Luengyosluechakul, S.; Pusoonthornthum, R.; Prapasarakul, N.; Kurilung, A.; Broens, E.M.; Wagenaar, J.A.; Goris, M.G.A.; Ahmed, A.A.; et al. Leptospira infection and shedding in dogs in Thailand. BMC Vet. Res. 2020, 16, 89. [Google Scholar] [CrossRef] [PubMed]
- Gay, N.; Soupé-Gilbert, M.-E.; Goarant, C. Though not Reservoirs, Dogs might Transmit Leptospira in New Caledonia. Int. J. Environ. Res. Public Health 2014, 11, 4316–4325. [Google Scholar] [CrossRef] [PubMed]
- Sant’Anna, R.; Vieira, A.S.; Grapiglia, J.; Lilenbaum, W. High number of asymptomatic dogs as leptospiral carriers in an endemic area indicates a serious public health concern. Epidemiol. Infect. 2017, 145, 1852–1854. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, R.S.; Di Azevedo, M.I.N.; Borges, A.L.d.S.B.; Carvalho-Costa, F.A.; Martins, G.; Lilenbaum, W. Persistent High Leptospiral Shedding by Asymptomatic Dogs in Endemic Areas Triggers a Serious Public Health Concern. Animals 2021, 11, 937. [Google Scholar] [CrossRef]
- Spangler, D.; Kish, D.; Beigel, B.; Morgan, J.; Gruszynski, K.; Naikare, H.; Nahar, V.K.; Coarsey, M.D.; Verma, A. Leptospiral shedding and seropositivity in shelter dogs in the Cumberland Gap Region of Southeastern Appalachia. PLoS ONE 2020, 15, e0228038. [Google Scholar] [CrossRef]
- Smith, A.M.; Arruda, A.G.; Evason, M.D.; Weese, J.S.; Wittum, T.E.; Szlosek, D.; Stull, J.W. A cross-sectional study of environmental, dog, and human-related risk factors for positive canine leptospirosis PCR test results in the United States, 2009 to 2016. BMC Vet. Res. 2019, 15, 412. [Google Scholar] [CrossRef]
- Harkin, K.R.; Roshto, Y.M.; Sullivan, J.T.; Purvis, T.J.; Chengappa, M.M. Comparison of polymerase chain reaction assay, bacteriologic culture, and serologic testing in assessment of prevalence of urinary shedding of leptospires in dogs. J. Am. Vet. Med. Assoc. 2003, 222, 1230–1233. [Google Scholar] [CrossRef]
- Miotto, B.A.; Tozzi, B.F.; Penteado, M.D.S.; Guilloux, A.; Moreno, L.Z.; Heinemann, M.B.; Moreno, A.M.; Lilenbaum, W.; Hagiwara, M.K. Diagnosis of acute canine leptospirosis using multiple laboratory tests and characterization of the isolated strains. BMC Vet. Res. 2018, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Thiermann, A.B. Canine leptospirosis in Detroit. Am. J. Vet. Res. 1980, 41, 1659–1661. [Google Scholar]
- de Freitas, J.C.; da Silva, F.G.; de Oliveira, R.C.; Delbem, A.C.B.; Muller, E.E.; Alves, L.A.; Teles, P.S. Isolation of Leptospira spp from dogs, bovine and swine naturally infected. Cienc. Rural 2004, 34, 853–856. [Google Scholar] [CrossRef]
- Koizumi, N.; Muto, M.M.; Akachi, S.; Okano, S.; Yamamoto, S.; Horikawa, K.; Harada, S.; Funatsumaru, S.; Ohnishi, M. Molecular and serological investigation of Leptospira and leptospirosis in dogs in Japan. J. Med. Microbiol. 2013, 62, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Harkin, K.R.; Hays, M.P. Variable-number tandem-repeat analysis of leptospiral DNA isolated from canine urine samples molecularly confirmed to contain pathogenic leptospires. J. Am. Vet. Med Assoc. 2016, 249, 399–405. [Google Scholar] [CrossRef]
- Balboni, A.; Zamagni, S.; Bertasio, C.; Boniotti, M.B.; Troìa, R.; Battilani, M.; Dondi, F. Identification of Serogroups Australis and Icterohaemorrhagiae in Two Dogs with a Severe Form of Acute Leptospirosis in Italy. Pathogens 2020, 9, 351. [Google Scholar] [CrossRef]
- Bertasio, C.; Boniotti, M.B.; Lucchese, L.; Ceglie, L.; Bellinati, L.; Mazzucato, M.; Furlanello, T.; D’Incau, M.; Natale, A.K. Detection of New Leptospira Genotypes Infecting Symptomatic Dogs: Is a New Vaccine Formulation Needed? Pathogens 2020, 9, 484. [Google Scholar] [CrossRef]
- Guagliardo, S.A.J.; Iverson, S.A.; Reynolds, L.; Yaglom, H.; Venkat, H.; Galloway, R.; Levy, C.; Reindel, A.; Sylvester, T.; Kretschmer, M.; et al. Despite high-risk exposures, no evidence of zoonotic transmission during a canine outbreak of leptospirosis. Zoonoses Public Health 2019, 66, 223–231. [Google Scholar] [CrossRef]
- Mori, M.; Bourhy, P.; Le Guyader, M.; Van Esbroeck, M.; Djelouadji, Z.; Septfons, A.; Kodjo, A.; Picardeau, M. Pet rodents as possible risk for leptospirosis, Belgium and France, 2009 to 2016. Eurosurveillance 2017, 22, 16–00792. [Google Scholar] [CrossRef]
- Dzupova, O.; Smiskova, D.; Huzova, Z.; Benes, J. Leptospirosis contracted from pet rats. Klin. Mikrobiol. Infekc. Lek. 2012, 18, 156–159. [Google Scholar]
- Guerra, B.; Schneider, T.; Luge, E.; Draeger, A.; Moos, V.; Loddenkemper, C.; Jansen, A.; Nöckler, K. Detection and characterization of Leptospira interrogans isolates from pet rats belonging to a human immunodeficiency virus-positive patient with leptospirosis. J. Med. Microbiol. 2008, 57, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Nordholm, A.C.; Omland, L.H.; Villumsen, S.; Al-Subeihe, I.; Katzenstein, T.L. Leptospirosis meningitis transmission from a pet mouse: A case report. J. Med. Case Rep. 2019, 13, 362. [Google Scholar] [CrossRef] [PubMed]
- Baer, R.; Turnberg, W.; Yu, D.; Wohrle, R. Leptospirosis in a Small Animal Veterinarian: Reminder to Follow Standardized Infection Control Procedures. Zoonoses Public Health 2009, 57, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Gaudie, C.M.; Featherstone, C.A.; Phillips, W.S.; McNaught, R.; Rhodes, P.M.; Errington, J.; Fearnley, C.; Fenner, J.S.; Pritchard, G.C. Human Leptospira interrogans serogroup icterohaemorrhagiae infection (Weil’s disease) acquired from pet rats. Vet. Rec. 2008, 163, 599–600. [Google Scholar] [CrossRef] [PubMed]
- Rajapakse, S.; Weeratunga, P.; Niloofa, R.; Fernando, N.; de Silva, N.; Rodrigo, C.; Maduranga, S.; Nandasiri, N.; Premawansa, S.; Karunanayake, L.; et al. A Diagnostic Scoring Model for Leptospirosis in Resource Limited Settings. PLoS Negl. Trop. Dis. 2016, 10, e0004513. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.D.; Gravekamp, C.; Carrington, D.G.; van de Kemp, H.; Hartskeerl, R.A.; Edwards, C.N.; Everard, C.O.; Terpstra, W.J.; Levett, P.N. Evaluation of the polymerase chain reaction for early diagnosis of leptospirosis. J. Med. Microbiol. 1995, 43, 110–114. [Google Scholar] [CrossRef]
- Riediger, I.N.; Stoddard, R.A.; Ribeiro, G.; Nakatani, S.M.; Moreira, S.D.R.; Skraba, I.; Biondo, A.W.; Reis, M.G.; Hoffmaster, A.R.; Vinetz, J.M.; et al. Rapid, actionable diagnosis of urban epidemic leptospirosis using a pathogenic Leptospira lipL32-based real-time PCR assay. PLoS Negl. Trop. Dis. 2017, 11, e0005940. [Google Scholar] [CrossRef]
- Sykes, J.E.; Hartmann, K.; Lunn, K.F.; Moore, G.E.; Stoddard, R.A.; Goldstein, R.E. 2010 ACVIM Small Animal Consensus Statement on Leptospirosis: Diagnosis, Epidemiology, Treatment, and Prevention. J. Vet. Intern. Med. 2011, 25, 1–13. [Google Scholar] [CrossRef]
- Fraune, C.K.; Schweighauser, A.; Francey, T. Evaluation of the diagnostic value of serologic microagglutination testing and a polymerase chain reaction assay for diagnosis of acute leptospirosis in dogs in a referral center. J. Am. Vet. Med Assoc. 2013, 242, 1373–1380. [Google Scholar] [CrossRef] [PubMed]
- Barr, S.C.; McDonough, P.L.; Scipioni-Ball, R.L.; Starr, J.K. Serologic responses of dogs given a commercial vaccine against Leptospira interrogans serovar pomona and Leptospira kirschneriserovar grippotyphosa. Am. J. Vet. Res. 2005, 66, 1780–1784. [Google Scholar] [CrossRef]
- Martin, L.; Wiggans, K.; Wennogle, S.; Curtis, K.; Chandrashekar, R.; Lappin, M. Vaccine-Associated Leptospira Antibodies in Client-Owned Dogs. J. Vet. Intern. Med. 2014, 28, 789–792. [Google Scholar] [CrossRef]
- Kodjo, A.; Calleja, C.; Loenser, M.; Lin, D.; Lizer, J. A Rapid In-Clinic Test Detects Acute Leptospirosis in Dogs with High Sensitivity and Specificity. BioMed Res. Int. 2016, 2016, 3760191. [Google Scholar] [CrossRef]
- Curtis, K.M.; Foster, P.C.; Smith, P.S.; Monn, M.P.; Stillman, B.A.; Chandrashekar, R.; Lappin, M.R.; Goldstein, R.E. Performance of a Recombinant LipL32 Based Rapid In-clinic ELISA (SNAP® Lepto) for the Detection of Antibodies against Leptospira in Dogs. Int. J. App. Res. Vet. Med. 2015, 13, 182–189. [Google Scholar]
- Winzelberg, S.; Tasse, S.M.; Goldstein, R.E.; Chapman, P.S.; Benedict, A.G.; Mason, G.D.; Noble, S.J.; Curtis, K.; Chandrashekar, R. Evaluation of SNAP® Lepto in the diagnosis of leptospirosis infections in dogs: Twenty two clinical cases. Int. J. Appl. Res. Vet. Med. 2015, 13, 193–198. [Google Scholar]
- Lizer, J.; Grahlmann, M.; Hapke, H.; Velineni, S.; Lin, D.; Kohn, B. Evaluation of a rapid IgM detection test for diagnosis of acute leptospirosis in dogs. Vet. Rec. 2017, 180, 517–2016. [Google Scholar] [CrossRef]
- Troìa, R.; Balboni, A.; Zamagni, S.; Frigo, S.; Magna, L.; Perissinotto, L.; Battilani, M.; Dondi, F. Prospective evaluation of rapid point-of-care tests for the diagnosis of acute leptospirosis in dogs. Vet. J. 2018, 237, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ellis, W.; Little, T. Current Topics in Veterinary Medicine and Animal Sciences; Springer: Berlin, Germany, 1986. [Google Scholar]
- Ellis, W.; Songer, J.; Montgomery, J.; Cassells, J. Prevalence of Leptospira interrogans serovar hardjo in the genital and urinary tracts of non-pregnant cattle. Vet. Rec. 1986, 118, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Babudieri, B. Animal reservoirs of leptospires. Ann. N. Y. Acad. Sci. 1958, 70, 393–413. [Google Scholar] [CrossRef]
- Guitian, J.; Thurmond, M.C.; Hietala, S.K. Infertility and abortion among first-lactation dairy cows seropositive or seronegative for Leptospira interrogans serovar Hardjo. J. Am. Vet. Med. Assoc. 1999, 215, 515–518. [Google Scholar] [PubMed]
- Steven, E.W.; Rogers, G.M.; Ramachandran, S.; Engelken, T.J.; Epperson, W.B.; Larson, R.L.; Maas, J.; Richey, E.; Bolin, C. Herd prevalence and risk factors of Leptospira infection in beef cow/calf operations in the United States. Bov. Pract. 2007, 41, 15–23. [Google Scholar]
- Ellis, W.; O'brien, J.; Neill, S.; Hanna, J. Bovine leptospirosis: Serological findings in aborting cows. Vet. Rec. 1982, 110, 178–180. [Google Scholar] [CrossRef]
- Arent, Z.; Frizzell, C.; Gilmore, C.; Allen, A.; Ellis, W. Leptospira interrogans serovars Bratislava and Muenchen animal infections: Implications for epidemiology and control. Vet. Microbiol. 2016, 190, 19–26. [Google Scholar] [CrossRef]
- Arent, Z.J.; Andrews, S.; Adamama-Moraitou, K.; Gilmore, C.; Pardali, D.; Ellis, W.A. Emergence of novel Leptospira serovars: A need for adjusting vaccination policies for dogs? Epidemiol. Infect. 2012, 141, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, N.; Nakajima, C.; Harunari, T.; Tanikawa, T.; Tokiwa, T.; Uchimura, E.; Furuya, T.; Mingala, C.N.; Villanueva, M.A.; Ohnishi, M.; et al. A New Loop-Mediated Isothermal Amplification Method for Rapid, Simple, and Sensitive Detection of Leptospira spp. in Urine. J. Clin. Microbiol. 2012, 50, 2072–2074. [Google Scholar] [CrossRef] [PubMed]
- Sonthayanon, P.; Limmathurotsakul, D.; Day, N.P.; Chierakul, W.; Peacock, S.J.; Wuthiekanun, V.; Thaipadungpanit, J.; Kalambaheti, T.; Boonsilp, S.; Amornchai, P.; et al. Accuracy of Loop-Mediated Isothermal Amplification for Diagnosis of Human Leptospirosis in Thailand. Am. J. Trop. Med. Hyg. 2011, 84, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Najian, A.N.; Syafirah, E.E.N.; Ismail, N.; Mohamed, M.; Yean, C.Y. Development of multiplex loop mediated isothermal amplification (m-LAMP) label-based gold nanoparticles lateral flow dipstick biosensor for detection of pathogenic Leptospira. Anal. Chim. Acta 2016, 903, 142–148. [Google Scholar] [CrossRef]
- Brogan, D.J.; Chaverra-Rodriguez, D.; Lin, C.P.; Smidler, A.L.; Yang, T.; Alcantara, L.M.; Antoshechkin, I.; Liu, J.; Raban, R.R.; Belda-Ferre, P.; et al. A Sensitive, Rapid, and Portable CasRx-based Diagnostic Assay for SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- de Puig, H.; Lee, R.A.; Najjar, D.; Tan, X.; Soenksen, L.R.; Angenent-Mari, N.M.; Donghia, N.M.; Weckman, N.E.; Ory, A.; Ng, C.F.; et al. Minimally instrumented SHERLOCK (miSHERLOCK) for CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging variants. Sci. Adv. 2021, 7, eabh2944. [Google Scholar] [CrossRef] [PubMed]
- Jirawannaporn, S.; Limothai, U.; Tachaboon, S.; Dinhuzen, J.; Kiatamornrak, P.; Chaisuriyong, W.; Bhumitrakul, J.; Mayuramart, O.; Payungporn, S.; Srisawat, N. Rapid and sensitive point-of-care detection of Leptospira by RPA-CRISPR/Cas12a targeting lipL32. PLoS Negl. Trop. Dis. 2022, 16, e0010112. [Google Scholar] [CrossRef]

| Assay | Specimen Type | Target | Comments |
|---|---|---|---|
| Darkfield microscopy | Urine | Leptospira organisms | Low sensitivity and specificity. Requires considerable technical expertise to interpret correctly. |
| Culture | Whole blood, urine | Leptospires | Special media required. Although sensitivity has historically been considered low and prolonged incubation times have been required, recent improvements in media have been associated with increased yields and shorter incubation times. |
| Microscopic agglutination test | Serum | Antibodies against various leptospiral serovars | False negatives can occur early in the course of illness or with immunosuppression, or when panels are used with limited numbers of serovars. False positives can occur with a history of vaccination in animals or with previous exposure. Paired titers performed at the same laboratory generally required for diagnosis. Inter-laboratory variation in results may occur. |
| Rapid diagnostic chromatographic or ELISA based tests | Serum or plasma | IgM or IgG against Leptospira | False negatives can occur early in the course of illness or with immunosuppression. False positives can occur with a history of vaccination in animals or with previous exposure. Weak positive results can be difficult to read. No information in infecting serogroup. |
| Histopathology | Kidney tissue collected via biopsy or necropsy | Leptospires | Organisms may be visualized with silver stains, immunohistochemistry, or fluorescence in situ hybridization. Antimicrobial therapy may lead to false-negative results. |
| Nucleic acid amplification tests | Blood, urine, CSF, tissue specimens | Leptospira DNA | Sensitivity and specificity unclear and may vary between assays offered by different laboratories. Antimicrobial therapy may lead to negative results. A positive result from a urine specimen may not have etiologic predictive value because of the potential for subclinical carriage. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sykes, J.E.; Reagan, K.L.; Nally, J.E.; Galloway, R.L.; Haake, D.A. Role of Diagnostics in Epidemiology, Management, Surveillance, and Control of Leptospirosis. Pathogens 2022, 11, 395. https://doi.org/10.3390/pathogens11040395
Sykes JE, Reagan KL, Nally JE, Galloway RL, Haake DA. Role of Diagnostics in Epidemiology, Management, Surveillance, and Control of Leptospirosis. Pathogens. 2022; 11(4):395. https://doi.org/10.3390/pathogens11040395
Chicago/Turabian StyleSykes, Jane E., Krystle L. Reagan, Jarlath E. Nally, Renee L. Galloway, and David A. Haake. 2022. "Role of Diagnostics in Epidemiology, Management, Surveillance, and Control of Leptospirosis" Pathogens 11, no. 4: 395. https://doi.org/10.3390/pathogens11040395
APA StyleSykes, J. E., Reagan, K. L., Nally, J. E., Galloway, R. L., & Haake, D. A. (2022). Role of Diagnostics in Epidemiology, Management, Surveillance, and Control of Leptospirosis. Pathogens, 11(4), 395. https://doi.org/10.3390/pathogens11040395







