SsNEP2 Contributes to the Virulence of Sclerotinia sclerotiorum
Abstract
:1. Introduction
2. Results
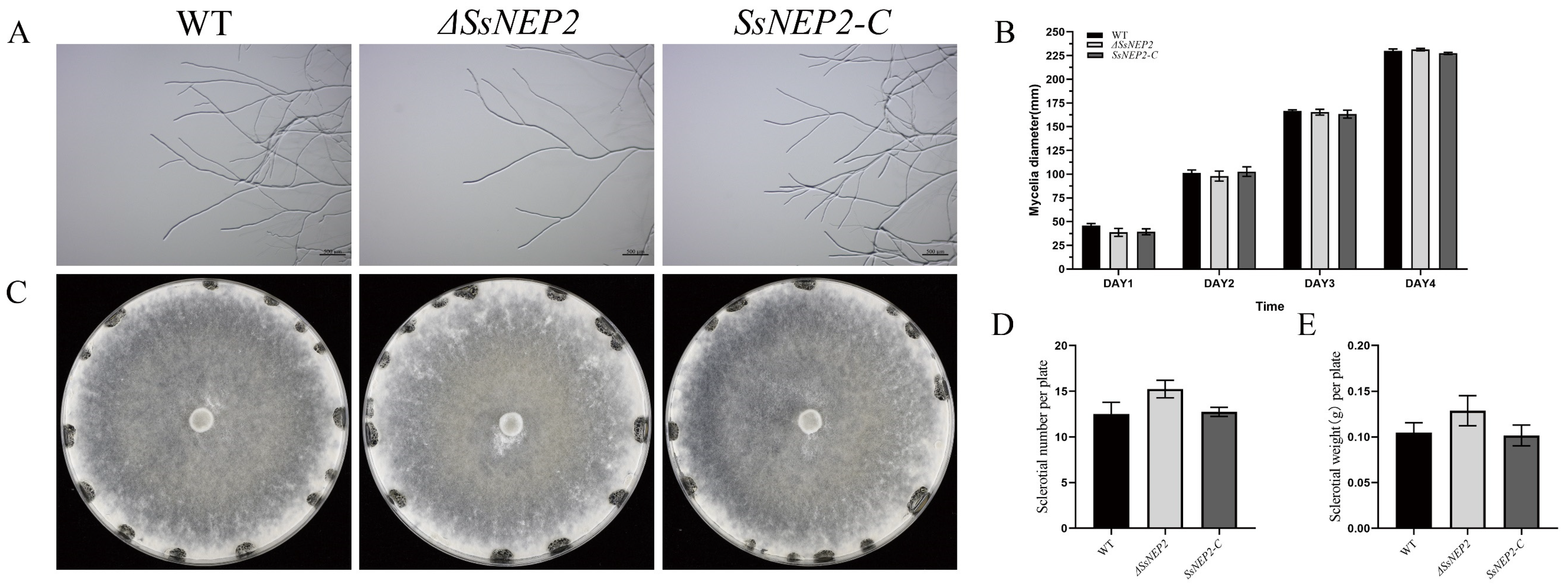
2.1. Knockout and Complementation of SsNEP2 in S. sclerotiorum
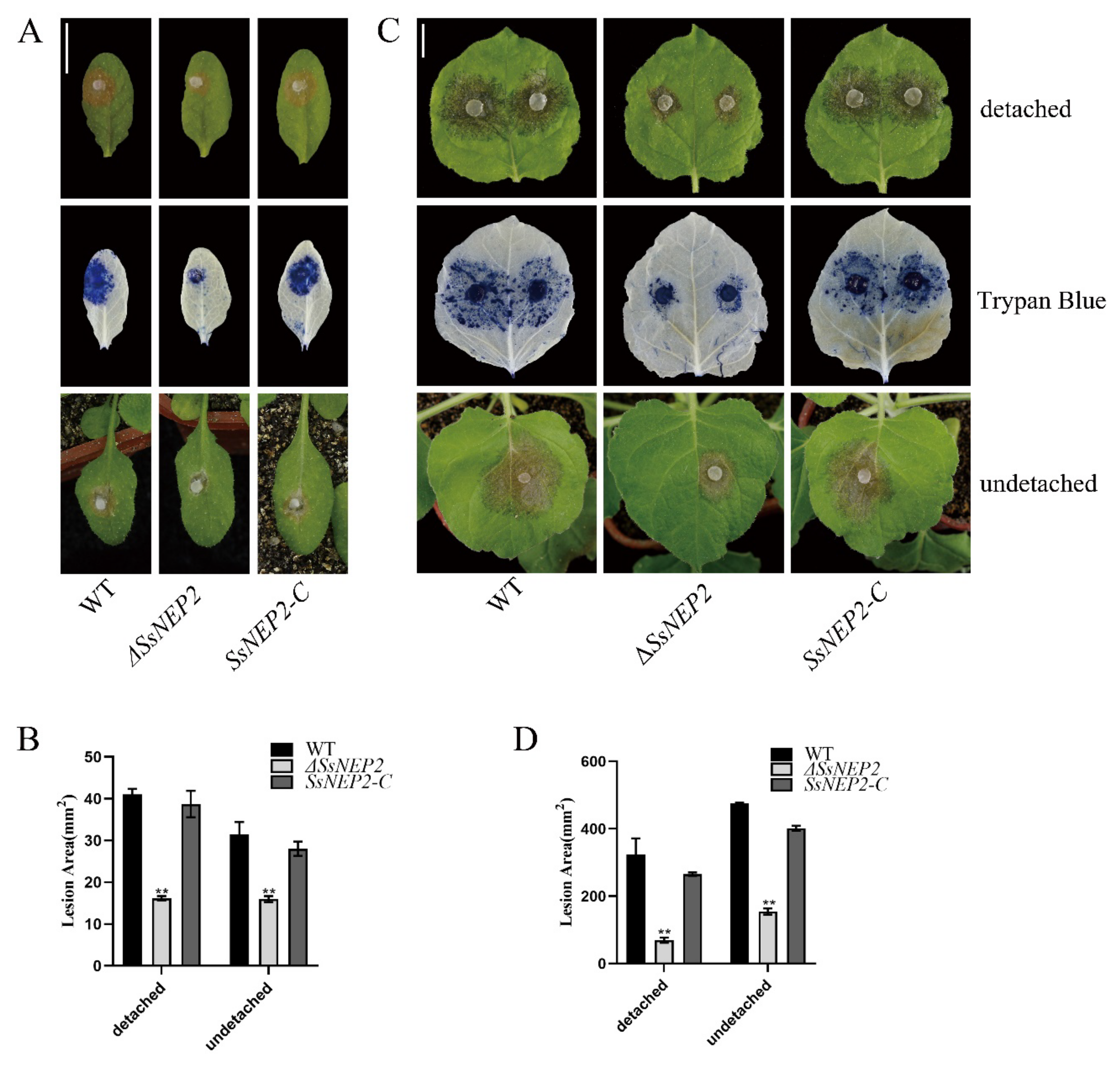
2.2. Loss of SsNEP2 Compromises Virulence of S. sclerotiorum
2.3. SsNEP2 Does Not Affect Oxalate Secretion and Appressorium Formation
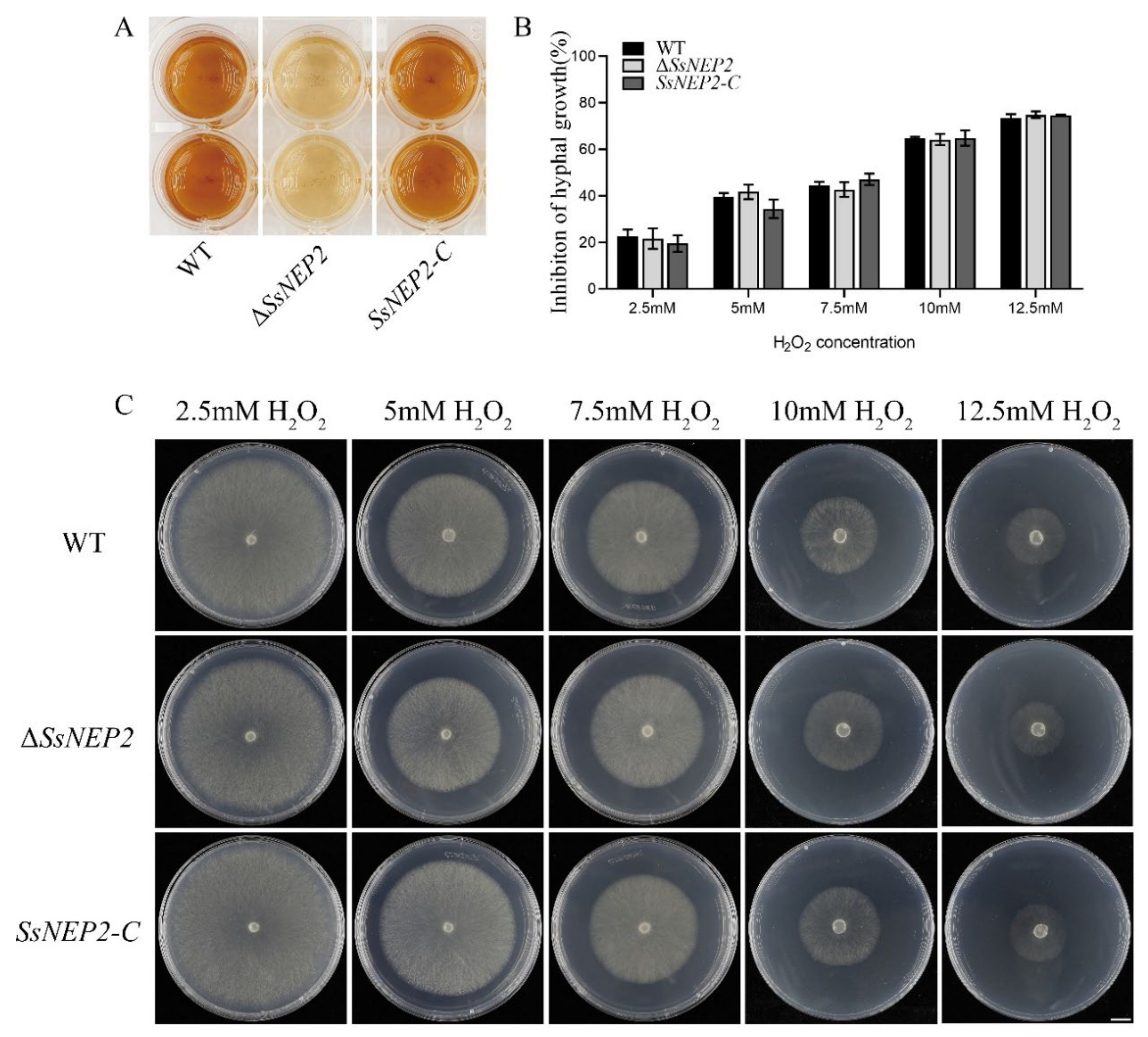
2.4. Knockout of SsNEP2 Affects ROS Accumulation
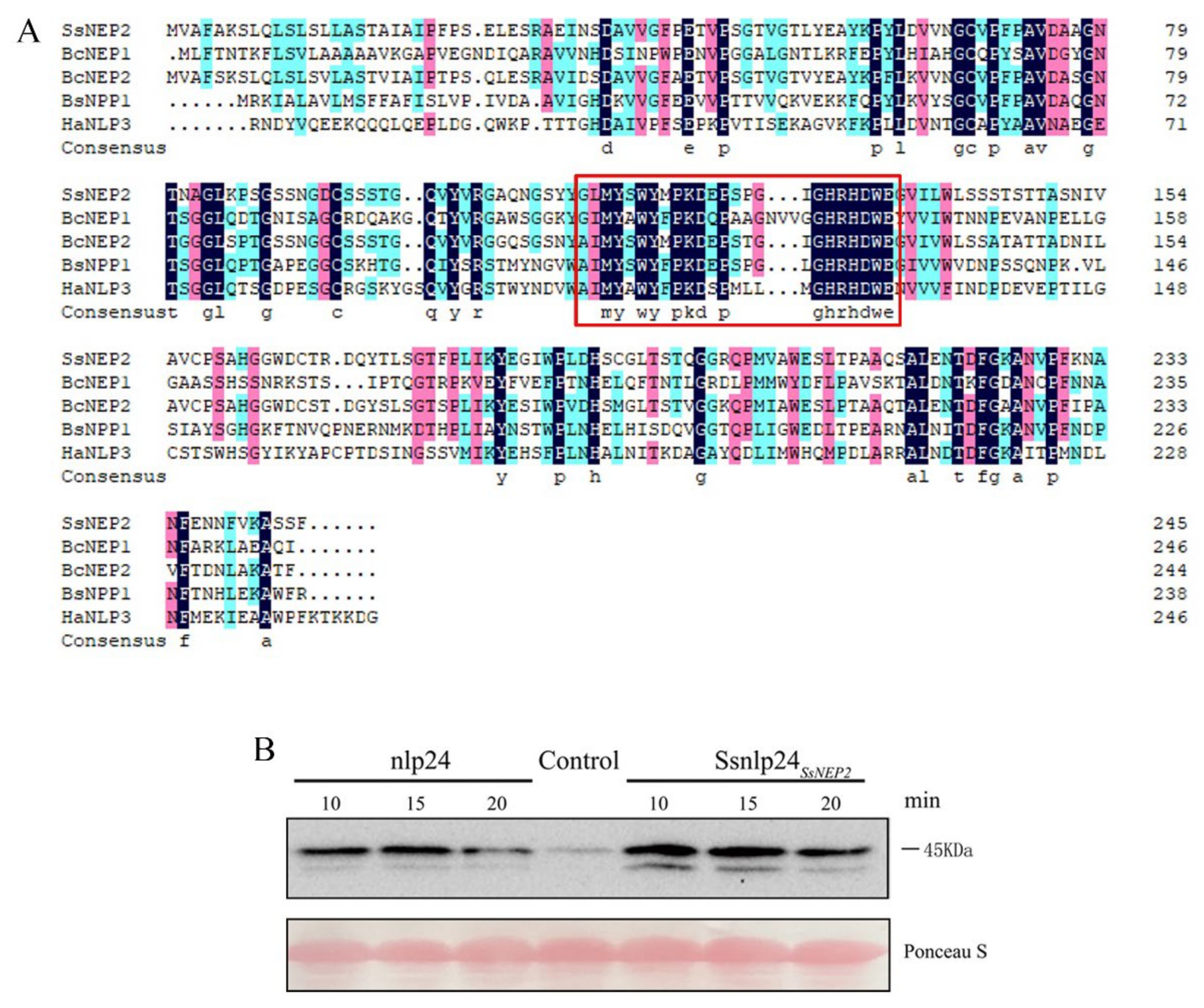
2.5. Ssnlp24 Derived from SsNEP2 Triggers MAPK Activation in Arabidopsis
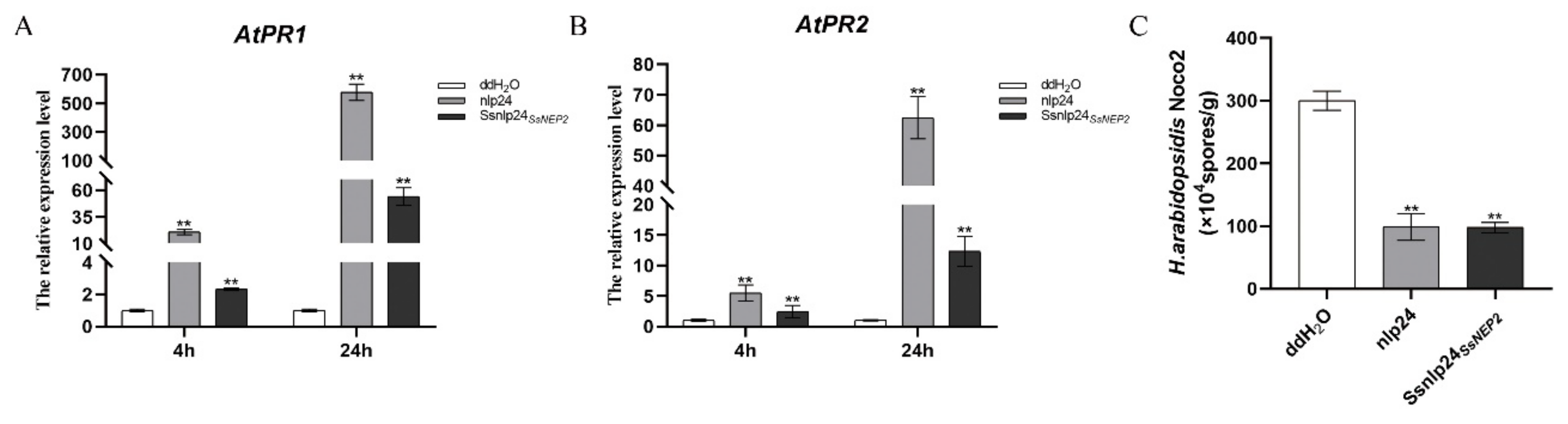
2.6. Ssnlp24SsNEP2 Acts as PAMP Signal to Induce Plant Immunity
3. Discussion
4. Materials and Methods
4.1. Fungal Strains and Culture Conditions
4.2. Plant Materials and Growth Conditions
4.3. Identification and Sequence Analysis of SsNEP2
4.4. Gene Replacement and Complementation
4.5. RT-qPCR Analysis
4.6. Analysis of Virulence
4.7. Compound Appressoria Observation and OA Analysis
4.8. DAB Staining
4.9. Pathogen Infection Assay
4.10. MAPK Activity Assay
4.11. Oxidative Stress Response
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, J.; Meng, J. Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2003, 106, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Xu, Y.; Hoy, R.; Zhang, J.; Qin, L.; Li, X. The Notorious Soilborne Pathogenic Fungus Sclerotinia sclerotiorum: An Update on Genes Studied with Mutant Analysis. Pathogens 2019, 9, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, X.; Rollins, J.A. Mechanisms of Broad Host Range Necrotrophic Pathogenesis in Sclerotinia sclerotiorum. Phytopathology 2018, 108, 1128–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, Q.; Cao, S.; Zhao, T.; Chen, L.; Zhuang, P.; Zhou, X.; Gao, Z. A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 2014, 86, 495–511. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xiao, J.; Du, J.; Yang, Y.; Bi, C.; Qing, L. Disruption of the Gene Encoding Endo-beta-1, 4-Xylanase Affects the Growth and Virulence of Sclerotinia sclerotiorum. Front. Microbiol. 2016, 7, 1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, C.; Dornenburg, H. Perspectives in the biological function and the technological application of polygalacturonases. Appl. Microbiol. Biotechnol. 2000, 53, 366–375. [Google Scholar] [CrossRef]
- Dallal Bashi, Z.; Rimmer, S.R.; Khachatourians, G.G.; Hegedus, D.D. Factors governing the regulation of Sclerotinia sclerotiorum cutinase A and polygalacturonase 1 during different stages of infection. Can. J. Microbiol. 2012, 58, 605–616. [Google Scholar] [CrossRef]
- Godoy, G.; Steadman, J.R.; Dickman, M.B.; Dam, R. Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 1990, 37, 179–191. [Google Scholar] [CrossRef]
- Williams, B.; Kabbage, M.; Kim, H.J.; Britt, R.; Dickman, M.B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 2011, 7, e1002107. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.S.; Min, J.Y.; Dickman, M.B. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe. Interact. 2008, 21, 605–612. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.S.; Li, G.Q.; Jiang, D.H.; Chen, W.D. Sclerotinia sclerotiorum: An Evaluation of Virulence Theories. Annu. Rev. Phytopathol. 2018, 56, 311–338. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Liberti, D.; Li, M.; Kim, Y.T.; Hutchens, A.; Wilson, R.; Rollins, J.A. Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 2015, 16, 559–571. [Google Scholar] [CrossRef] [PubMed]
- Varden, F.A.; De la Concepcion, J.C.; Maidment, J.H.; Banfield, M.J. Taking the stage: Effectors in the spotlight. Curr. Opin Plant Biol. 2017, 38, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yang, G.; Ma, M.; Liu, X.; Li, B.; Xie, J.; Fu, Y.; Chen, T.; Yu, Y.; Chen, W.; et al. An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 2020, 21, 686–701. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Tang, L.; Gong, Y.; Xie, J.; Fu, Y.; Jiang, D.; Li, G.; Collinge, D.B.; Chen, W.; Cheng, J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755. [Google Scholar] [CrossRef] [Green Version]
- Pemberton, C.L.; Salmond, G.P. The Nep1-like proteins-a growing family of microbial elicitors of plant necrosis. Mol. Plant Pathol. 2004, 5, 353–359. [Google Scholar] [CrossRef]
- Bailey, B.A. Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology 1995, 85, 1250–1255. [Google Scholar] [CrossRef]
- Bailey, B.A.; Apel-Birkhold, P.C.; Luster, D.G. Expression of NEP1 by Fusarium oxysporum f. sp. erythroxyli After Gene Replacement and Overexpression Using Polyethylene Glycol-Mediated Transformation. Phytopathology 2002, 92, 833–841. [Google Scholar] [CrossRef] [Green Version]
- Ottmann, C.; Luberacki, B.; Kufner, I.; Koch, W.; Brunner, F.; Weyand, M.; Mattinen, L.; Pirhonen, M.; Anderluh, G.; Seitz, H.U.; et al. A common toxin fold mediates microbial attack and plant defense. Proc. Natl. Acad. Sci. USA 2009, 106, 10359–10364. [Google Scholar] [CrossRef] [Green Version]
- Fellbrich, G.; Romanski, A.; Varet, A.; Blume, B.; Brunner, F.; Engelhardt, S.; Felix, G.; Kemmerling, B.; Krzymowska, M.; Nurnberger, T. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002, 32, 375–390. [Google Scholar] [CrossRef]
- Seidl, M.F.; Van den Ackerveken, G. Activity and Phylogenetics of the Broadly Occurring Family of Microbial Nep1-Like Proteins. Annu. Rev. Phytopathol. 2019, 57, 367–386. [Google Scholar] [CrossRef] [PubMed]
- Lenarcic, T.; Albert, I.; Bohm, H.; Hodnik, V.; Pirc, K.; Zavec, A.B.; Podobnik, M.; Pahovnik, D.; Zagar, E.; Pruitt, R.; et al. Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 2017, 358, 1431–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santhanam, P.; van Esse, H.P.; Albert, I.; Faino, L.; Nurnberger, T.; Thomma, B.P. Evidence for functional diversification within a fungal NEP1-like protein family. Mol. Plant Microbe Interact. 2013, 26, 278–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattinen, L.; Tshuikina, M.; Mae, A.; Pirhonen, M. Identification and characterization of Nip, necrosis-inducing virulence protein of Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 2004, 17, 1366–1375. [Google Scholar] [CrossRef] [Green Version]
- Pemberton, C.L.; Whitehead, N.A.; Sebaihia, M.; Bell, K.S.; Hyman, L.J.; Harris, S.J.; Matlin, A.J.; Robson, N.D.; Birch, P.R.; Carr, J.P.; et al. Novel quorum-sensing-controlled genes in Erwinia carotovora subsp. carotovora: Identification of a fungal elicitor homologue in a soft-rotting bacterium. Mol. Plant Microbe Interact. 2005, 18, 343–353. [Google Scholar] [CrossRef] [Green Version]
- Albert, I.; Bohm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 2015, 1, 15140. [Google Scholar] [CrossRef]
- Bohm, H.; Albert, I.; Oome, S.; Raaymakers, T.M.; Van den Ackerveken, G.; Nurnberger, T. A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 2014, 10, e1004491. [Google Scholar] [CrossRef]
- Ono, E.; Mise, K.; Takano, Y. RLP23 is required for Arabidopsis immunity against the grey mould pathogen Botrytis cinerea. Sci. Rep. 2020, 10, 13798. [Google Scholar] [CrossRef]
- Oome, S.; Raaymakers, T.M.; Cabral, A.; Samwel, S.; Bohm, H.; Albert, I.; Nurnberger, T.; Van den Ackerveken, G. Nep1-like proteins from three kingdoms of life act as a microbe-associated molecular pattern in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 16955–16960. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Nie, J.; Chang, Y.; Huang, L. Nep1-like Proteins from Valsa mali Differentially Regulate Pathogen Virulence and Response to Abiotic Stresses. J. Fungi 2021, 7, 830. [Google Scholar] [CrossRef]
- Dallal Bashi, Z.; Hegedus, D.D.; Buchwaldt, L.; Rimmer, S.R.; Borhan, M.H. Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Mol. Plant Pathol. 2010, 11, 43–53. [Google Scholar] [CrossRef]
- Xu, L.; Xiang, M.; White, D.; Chen, W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol 2015, 17, 2896–2909. [Google Scholar] [CrossRef]
- Kabbage, M.; Yarden, O.; Dickman, M.B. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 2015, 233, 53–60. [Google Scholar] [CrossRef]
- Jiao, W.; Yu, H.; Cong, J.; Xiao, K.; Zhang, X.; Liu, J.; Zhang, Y.; Pan, H. Transcription factor SsFoxE3 activating SsAtg8 is critical for sclerotia, compound appressoria formation, and pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2022, 23, 204–217. [Google Scholar] [CrossRef]
- Schouten, A.; Van Baarlen, P.; Van Kan, J.A.L. Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associate with membranes and the nucleus of plant cells. New Phytol. 2008, 177, 493–505. [Google Scholar] [CrossRef]
- Qin, G.; Tian, S.; Chan, Z.; Li, B. Crucial role of antioxidant proteins and hydrolytic enzymes in pathogenicity of Penicillium expansum: Analysis based on proteomics approach. Mol. Cell Proteom. 2007, 6, 425–438. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Willger, S.D.; Park, S.W.; Puttikamonkul, S.; Grahl, N.; Cho, Y.; Mukhopadhyay, B.; Cramer, R.A., Jr.; Lawrence, C.B. TmpL, a transmembrane protein required for intracellular redox homeostasis and virulence in a plant and an animal fungal pathogen. PLoS Pathog. 2009, 5, e1000653. [Google Scholar] [CrossRef] [Green Version]
- Ding, L.N.; Li, T.; Guo, X.J.; Li, M.; Liu, X.Y.; Cao, J.; Tan, X.L. Sclerotinia Stem Rot Resistance in Rapeseed: Recent Progress and Future Prospects. J. Agric. Food Chem. 2021, 69, 2965–2978. [Google Scholar] [CrossRef]
- Pirc, K.; Hodnik, V.; Snoj, T.; Lenarcic, T.; Caserman, S.; Podobnik, M.; Bohm, H.; Albert, I.; Kotar, A.; Plavec, J.; et al. Nep1-like proteins as a target for plant pathogen control. PLoS Pathog. 2021, 17, e1009477. [Google Scholar] [CrossRef]
- Lehmann, S.; Serrano, M.; L’Haridon, F.; Tjamos, S.E.; Metraux, J.P. Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry 2015, 112, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Wang, Q.; Sun, Y.; Zhang, Y.; Zhang, X.; Liu, J.; Yu, G.; Pan, H. Sssfh1, a Gene Encoding a Putative Component of the RSC Chromatin Remodeling Complex, Is Involved in Hyphal Growth, Reactive Oxygen Species Accumulation, and Pathogenicity in Sclerotinia sclerotiorum. Front. Microbiol. 2018, 9, 1828. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, D.; Chen, T.; Li, B.; Zhang, Z.; Qin, G.; Tian, S. Production, Signaling, and Scavenging Mechanisms of Reactive Oxygen Species in Fruit-Pathogen Interactions. Int. J. Mol. Sci. 2019, 20, 2994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Chen, C.; Kabbage, M.; Dickman, M.B. Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl. Environ. Microbiol. 2011, 77, 7721–7729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heller, J.; Tudzynski, P. Reactive oxygen species in phytopathogenic fungi: Signaling, development, and disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef]
- Rollins, J.A. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. 2003, 16, 785–795. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Ao, K.; Tian, L.; Qiu, Y.; Huang, X.; Liu, X.; Hoy, R.; Zhang, Y.; Rashid, K.Y.; Xia, S.; et al. A Forward Genetic Screen in Sclerotinia sclerotiorum Revealed the Transcriptional Regulation of Its Sclerotial Melanization Pathway. Mol. Plant Microbe Interact. 2022, 35, MPMI10210254R. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Li, W.; Huang, X.; Tang, X.; Qin, L.; Liu, Y.; Xia, Y.; Peng, Z.; Xia, S. SsNEP2 Contributes to the Virulence of Sclerotinia sclerotiorum. Pathogens 2022, 11, 446. https://doi.org/10.3390/pathogens11040446
Yang C, Li W, Huang X, Tang X, Qin L, Liu Y, Xia Y, Peng Z, Xia S. SsNEP2 Contributes to the Virulence of Sclerotinia sclerotiorum. Pathogens. 2022; 11(4):446. https://doi.org/10.3390/pathogens11040446
Chicago/Turabian StyleYang, Chenghuizi, Wei Li, Xingchuan Huang, Xianyu Tang, Lei Qin, Yanan Liu, Yunong Xia, Zhihong Peng, and Shitou Xia. 2022. "SsNEP2 Contributes to the Virulence of Sclerotinia sclerotiorum" Pathogens 11, no. 4: 446. https://doi.org/10.3390/pathogens11040446





