Multidrug-Resistant and Extended-Spectrum β-Lactamase (ESBL) - Producing Enterobacterales Isolated from Carriage Samples among HIV Infected Women in Yaoundé, Cameroon
Abstract
1. Introduction
2. Results
2.1. Sociodemographic and Clinical Characteristics
2.2. Distribution of Bacterial Species According to Multidrug Resistance and ESBL Production
2.3. Antibiotic Resistance Patterns and Distribution of Resistances Mechanism among MDR-Enterobacterales
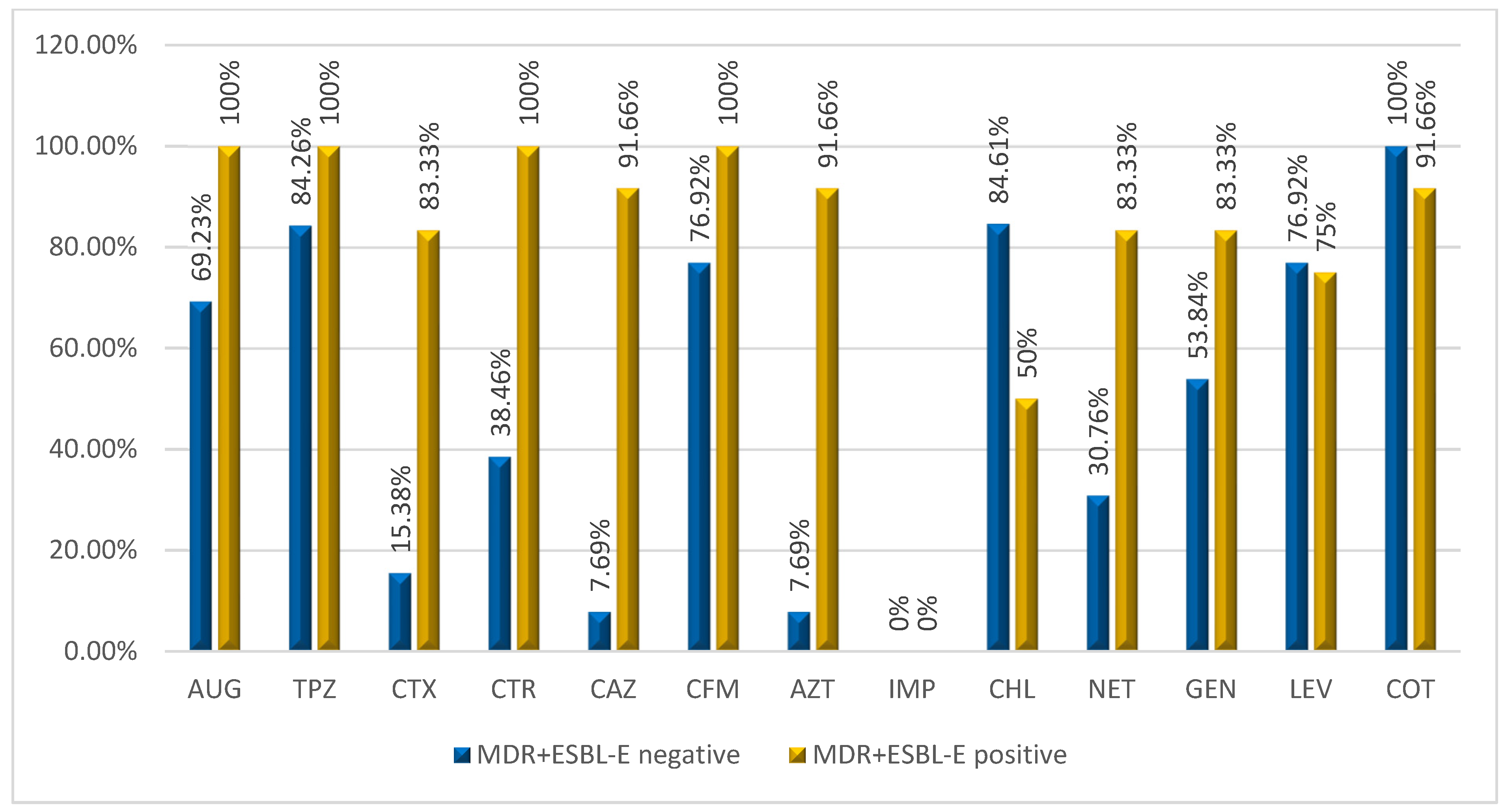
2.3.1. Antibiotic Resistance among MDR-Enterobacterales Isolated
2.3.2. Resistance Mechanisms among MDR -Enterobacterales isolated in WLHIV at Yaoundé Central Hospital
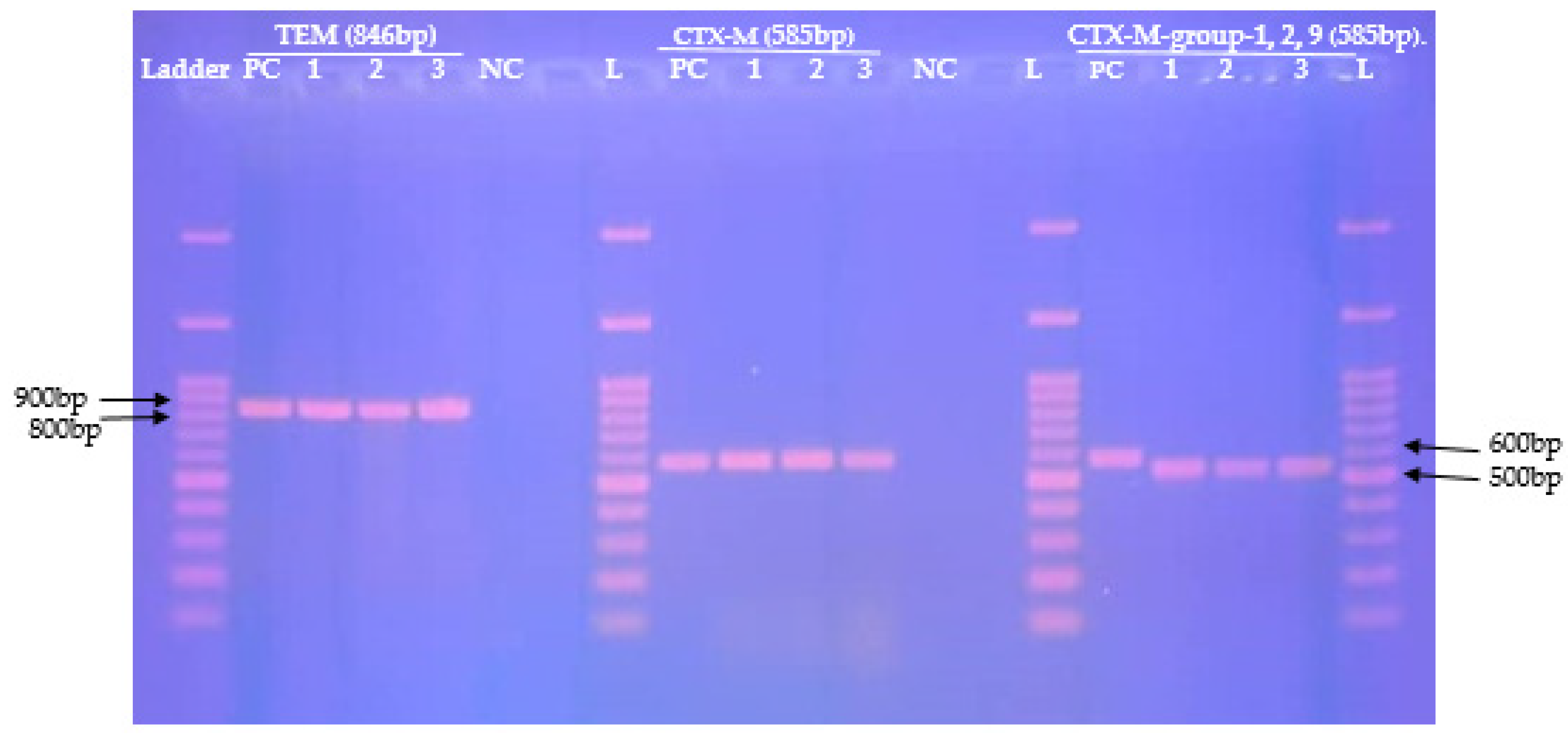
2.4. Distribution of β-Lactamase Genes Mediating Resistance in MDR-Enterobacterales
3. Discussion
4. Materials and Methods
4.1. Study Settings
4.2. Study Population
4.3. Sample Collection
4.4. Laboratory Analyses
4.4.1. Culture and Identification
4.4.2. Antimicrobial Susceptibility Testing (AST)
4.4.3. Screening and Confirmation for ESBL- and Carbapenemase -Producing Enterobacterales
4.4.4. Genomic Extraction
4.4.5. Conventional Polymerase Chain Reaction
4.4.6. DNA Electrophoresis and Visualization
4.5. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gandra, S.; Alvarez-Uria, G.; Turner, P.; Joshi, J.; Limmathurotsakul, D.; van Doorn, H.R. Antimicrobial Resistance Surveillance in Low- and Middle-Income Countries: Progress and Challenges in Eight South Asian and Southeast Asian Countries. Clin. Microbiol. Rev. 2020, 33, 1–29s. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L. 313-Infections Due to Other Members of the Enterobacteriaceae, Including Management of Multidrug-Resistant Strains. In Goldman’s Cecil Medicine, 24th ed.; Goldman, L., Schafer, A.I., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2012; Volume 2, pp. 874–877. [Google Scholar]
- WHO. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017; pp. 1–88. [Google Scholar]
- Baux-Pomarès, E. Traitement Des Infections À Entérobactéries Sécrétrices De BLSE: Alternatives Aux Carbapénèmes. Ph.D. Thesis, Université de Lorraine, Metz, France, 2015. [Google Scholar]
- Wilmore, S.M.S.; Kranzer, K.; Williams, A.; Makamure, B.; Nhidza, A.F.; Mayini, J.; Bandason, T.; Metcalfe, J.; Nicol, M.P.; Balakrishnan, I.; et al. Carriage of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Hiv-Infected Children in Zimbabwe. J. Med. Microbiol. 2017, 66, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Manyahi, J.; Moyo, S.J.; Tellevik, M.G.; Langeland, N.; Blomberg, B. High Prevalence of Fecal Carriage of Extended Spectrum Beta-Lactamase-Producing Enterobacteriaceae among Newly Hiv-Diagnosed Adults in a Community Setting in Tanzania. Microb. Drug Resist. 2020, 26, 1540–1545. [Google Scholar] [CrossRef] [PubMed]
- Mandal, D.K.; Sah, S.K.; Mishra, S.K.; Sharma, S.; Kattel, H.P.; Pandit, S.; Yadav, P.K.; Laghu, U.; Lama, R.; Sah, N.P.; et al. Carriage of Extended-Spectrum-Beta-Lactamase- and Ampc-Beta-Lactamase-Producing Enterobacteriaceae (Esbl-Pe) in Healthy Community and Outpatient Department (Opd) Patients in Nepal. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, e0264818. [Google Scholar] [CrossRef] [PubMed]
- ONUSIDA. Estimations Épidémiologiques Préliminaires De L’onusida 2021. Stat. Mond. Sur Vih 2021, 2021, 1–7. [Google Scholar]
- Institut National De La Statistique (Ins) Et Icf. International. Enquête Démographique et de Santé et à Indicateurs Multiples du Cameroun 2018; INS et ICF International: Yaoundé, Cameroun; Rockville, MD, USA, 2020; pp. 1–696. [Google Scholar]
- Bayleyegn, B.; Fisaha, R.; Kasew, D. Fecal Carriage of Extended Spectrum Beta-Lactamase Producing Enterobacteriaceae among Hiv Infected Children at the University of Gondar Comprehensive Specialized Hospital Gondar, Ethiopia. AIDS Res. Ther. 2021, 18, 19. [Google Scholar] [CrossRef] [PubMed]
- Teklu, D.S.; Negeri, A.A.; Legese, M.H.; Bedada, T.L.; Woldemariam, H.K.; Tullu, K.D. Extended-Spectrum Beta-Lactamase Production and Multi-Drug Resistance among Enterobacteriaceae Isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control 2019, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Toudji, A.G.; Djeri, B.; Karou, S.D.; Tigossou, S.; Ameyapoh, Y.; de Souza, C. Prévalence Des Souches D’entérobactéries Productrices De Bêta-Lactamases À Spectre Élargi Isolées Au Togo Et De Leur Sensibilité Aux Antibiotiques. Int. J. Biol. Chem. Sci. 2017, 11, 1165–1177. [Google Scholar] [CrossRef]
- Mabeku Kouitcheu, L.B.; Efon Ekangouo, A.; Kouam Mewa, J.E. Trends in Carriage of Extended Spectrum Β-Lactamase and Carbapenemase Producing Enterobacteria among Hiv Infected Patients and Their Antimicrobial Susceptibility Patterns in Western-Cameroon. Res. Sq. 2020, 1–20. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F. Dissemination of Carbapenemases (Kpc, Ndm, Oxa-48, Imp, and Vim) among Carbapenem-Resistant Enterobacteriaceae Isolated from Adult and Children Patients in China. Front. Cell Infect. Microbiol. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. Ampc Beta-Lactamases. Clin. Microbiol. 2009, 22, 61–82. [Google Scholar]
- Hosuru, S.S.; Bairy, I.; Nayak, N.; Padukone, S.; Sathian, B.; Gokhale, S. Low Rate of Gut Colonization by Extended-Spectrum Β-Lactamase Producing Enterobacteriaceae in Hiv Infected Persons as Compared to Healthy Individuals in Nepal. PLoS ONE 2019, 14, e0212042. [Google Scholar]
- Mohamed, E.S.; Khairy, R.M.M.; Abdelrahim, S.S. Prevalence and Molecular Characteristics of Esbl and Ampc Beta -Lactamase Producing Enterobacteriaceae Strains Isolated from Utis in Egypt. Antimicrob. Resist. Infect. Control 2020, 9, 198. [Google Scholar] [CrossRef] [PubMed]
- Lonchel, C.M.; Meex, C.; Gangoué-Piéboji, J.; Boreux, R.; Assoumou, M.C.; Melin, P.; De Mol, P. Proportion of Extended-Spectrum ß-Lactamase-Producing Enterobacteriaceae in Community Setting in Ngaoundere, Cameroon. BMC Infect. Dis. 2012, 12, 53. [Google Scholar] [CrossRef] [PubMed]
- Magoue, C.L.; Melin, P.; Gangoue-Pieboji, J.; Okomo Assoumou, M.C.; Boreux, R.; De Mol, P. Prevalence and Spread of Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Ngaoundere, Cameroon. Clin. Microbiol. Infect. 2013, 19, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Société Française de Microbiologie. CASFM/EUCAST 2021; Société Française de Microbiologie Ed: Paris, France, 2021; pp. 1–188. [Google Scholar]
- Ahmed, O.B.; Dablool, A.S. Quality Improvement of the DNA Extracted by Boiling Method in Gram Negative Bacteria. Int. J. Bioassays 2017, 6, 5347–5349. [Google Scholar] [CrossRef]
- Strauß, L.M.; Dahms, C. Development and Evaluation of a Novel Universal Beta-Lactamase Gene Sutyping Assy for Blatem, Blactx-M, Blactx-M1, 2, 9, Blashv Using Clinical and Livestock-Associated Esherichia coli. J. Antimicrob. Chemother. 2014, 70, 710–715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Batchelor, M.; Hopkins, K.; Threlfall, E.J.; Clifton-Hadley, F.A.; Stallwood, A.D.; Davies, R.H.; Liebana, E. Bla(Ctx-M) Genes in Clinical Salmonella Isolates Recovered from Humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 2005, 49, 1319–1322. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; Hamilton, N.; Church, D.L.; Nordmann, P.; Poirel, L. Development and Clinical Validation of a Molecular Diagnostic Assay to Detect Ctx-M-Type Beta-Lactamases in Enterobacteriaceae. Clin. Microbiol. Infect. 2007, 13, 291–297. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Variables | Enterobacterales Positive n (%) | MDR-Enterobacterales Positive n (%) | MDR-ESBL-Enterobacterales Positive n (%) |
|---|---|---|---|
| Overall | 37 (31.0) | 23 (62.0) | 11 (47.82) |
| Age | |||
| Median [IQR #] | 39 [34.0; 44.0] | 36 [31.0; 41.0] | 36 [31.0; 41.0] |
| Mean [std *] | 38.7 [7.3] | 36 [7.1] | 36 [7.1] |
| Age Group | |||
| 20–24 | 2 (5.41) | 1 (50.0) | 1 (50.0) |
| 25–29 | 2 (5.41) | 1 (50.0) | 1 (50.0) |
| 30–34 | 7 (18.92) | 2 (28.0) | 2 (28.57) |
| 35–39 | 10 (27.03) | 6 (60.0) | 2 (20.0) |
| 40–44 | 8 (21.62) | 7 (87.50) | 1 (12.50) |
| >45 | 8 (21.62) | 6 (75.0) | 3 (37.50) |
| Education | |||
| Illiterate | 0 | 0 | 0 |
| Primary school | 4 (10.81) | 3 (75.0) | 2 (50.0) |
| High school | 24 (64.86) | 16 (66.67) | 6 (25.0) |
| University | 9 (24.32) | 4 (44.44) | 2 (22.22) |
| Profession | |||
| Seller | 6 (16.22) | 5 (83.33) | 2 (33.33) |
| Student | 2 (5.41) | 1 (50.0) | 1 (50.0) |
| Housewife | 7 (18.92) | 3 (42.86) | 2 (28.57) |
| Civil servant | 8 (21.62) | 6 (75.0) | 3 (37.50) |
| Farmer | 13 (35.14) | 7 (53.85) | 2 (15.38) |
| Healthcare worker | 1 (2.70) | 1 (100) | 0 |
| Pregnancy Status | |||
| No | 32 (86.49) | 20 (62.50) | 9 (28.12) |
| Yes | 5 (13.51) | 3 (60.0) | 1 (20.0) |
| Vaginal Hygiene | |||
| No | 7 (18.92) | 4 (57.14) | 1 (14.29) |
| Yes | 30 (81.08) | 19 (63.33) | 9 (30.0) |
| Previous antibiotic | |||
| No | 0 | 0 | 0 |
| Yes | 37 (100) | 23 (62.16) | 10 (27.03) |
| Viral Load | |||
| Unknown | 5 (13.51) | 3 (60.0) | 3 (60.0) |
| <1000 copies | 1 (2.70) | 1 (100) | 0 |
| >1000 copies | 6 (16.22) | 4 (66.67) | 3 (50.0) |
| Undetectable | 25 (67.57) | 15 (60.0) | 4 (16.0) |
| Period under ARV Treatment | |||
| 1–5 years | 15 (40.54) | 8 (53.33) | 2 (13.33) |
| 6–10 years | 9 (24.32) | 7 (77.78) | 4 (44.44) |
| 10–15 years | 11 (29.73) | 7 (63.64) | 3 (27.27) |
| 16–20 years | 2 (5.41) | 1 (50.0) | 1 (50.0) |
| >20 years | 0 | 0 | 0 |
| Bacterial Species (n) | MDR-ESBL Negative n (%) | MDR+ESBL Negative n (%) | p-Value | MDR+ESBL Positive n (%) | p-Value |
|---|---|---|---|---|---|
| Escherichia coli (29) | 15 (51.72) | 9 (31.03) | <0.0001 | 5 (17.24) | <0.0001 |
| Klebsiella pneumoniae (7) | 2 (28.57) | 1 (14.28) | 4 (57.14) | ||
| Klebsiella oxytoca (2) | 0 | 1 (50.0) | 1 (50.0) | ||
| C. freundii (1) | 1 (100) | 0 | 0 | ||
| Enterobacter aerogenes (1) | 0 | 1 (100) | 0 (0) | ||
| Enterobacter sakazaki (1) | 1 (100) | 0 | 0 | ||
| Enterobacter gergoviae (2) | 1 (50.0) | 0 | 1 (50.0) | ||
| Serratia fonticola (2) | 1 (50.0) | 0 (0) | 1 (50.0) | ||
| Serratia marcescens (2) | 2 (100) | 0 | 0 | ||
| Raoultella ornithynolitica (1) | 0 | 1 (100) | 0 (0) | ||
| Overall, N = 48 | 23 (48.0) | 13 (27.0) | 12 (25.0) |
| Bacterial Species | ß-Lactams n (%) | ||||||
| AUG | TPZ | CTX | CTR | CAZ | AZT | IMP | |
| Overall (n = 25) | 22 (88.0) | 24 (96.0) | 11 (44.0) | 14 (56.0) | 14 (56.0) | 12 (48.0) | 0 |
| E. coli (n = 14) | 12 (85.7) | 14 (100) | 5 (35.71) | 6 (42.85) | 6 (42.85) | 5 (35.71) | 0 |
| K. pneumoniae (n = 5) | 4 (80.0) | 5 (100) | 4 (80.0) | 5 (100) | 5 (100) | 4 (80.0) | 0 |
| E. gergoviae (n = 1) | 1 (100) | 1 (100) | 0 | 1 (100) | 1 (100) | 1 (100) | 0 |
| E. aerogenes (n = 1) | 1 (100) | 1 (100) | 0 | 0 | 0 | 0 | 0 |
| S. fonticola (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 0 |
| K. oxytoca (n = 2) | 2 (100) | 2 (100) | 1 (50.0) | 1 (50.0) | 1 (50.0) | 1 (50.0) | 0 |
| R. ornithynolitica (n = 1) | 1 (100) | 0 | 0 | 0 | 0 | 0 | 0 |
| Bacterial Species | Non-ß-Lactam Antibiotics n (%) | ||||||
| GEN | NET | TMP/SXT | CHL | LEV | |||
| Overall (n = 25) | 18 (72.0) | 14 (56.0) | 24 (96.0) | 18 (72.0) | 20 (80.0) | ||
| E. coli (n = 14) | 10 (71.42) | 6 (42.85) | 14 (100) | 11 (78.57) | 10 (71.42) | ||
| K. pneumoniae (n = 5) | 4 (80.0) | 3 (60.0) | 4 (80.0) | 2 (50.0) | 4 (80.0) | ||
| E. gergoviae (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ||
| E. aerogenes (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ||
| S. fonticola (n = 1) | 1 (100) | 1 (100) | 1 (100) | 0 | 1 (100) | ||
| K. oxytoca (n = 2) | 1 (50.0) | 1 (50.0) | 2 (100) | 2 (100) | 2 (100) | ||
| R. ornithynolitica (n = 1) | 0 | 1 (100) | 1 (100) | 1 (100) | 1 (100) | ||
| Phenotypic Profile | Number of Family of Antibiotics | Number of Antibiotics | Number of Isolates (%) |
|---|---|---|---|
| TPZ-CHL-LEV-COT | 4 | 4 | 2 (8.33) |
| AUG-TPZ-NET-CHL-COT | 4 | 5 | 1 (4.16) |
| AUG-TPZ-GEN-CHL-COT | 2 (8.33) | ||
| AUG-TPZ-CHL-LEV-COT | 1 (4.16) | ||
| AUG-TPZ-GEN-CHL-LEV-COT | 5 | 6 | 2 (8.33) |
| AUG-TPZ-AZT-GEN-CHL-COT | 5 | 1 (416) | |
| AUG-TPZ-NET-GEN-CHL-COT | 4 | 1 (4.16) | |
| AUG-CTR-NET-CHL-LEV-COT | 6 | 1 (4.16) | |
| CTX-CTR-CAZ-ATM-CHL-COT | 4 | 6 | 1 (4.16) |
| AUG-TPZ-CTR-CAZ-AZT-NET-COT-LEV | 6 | 8 | 1 (4.16) |
| AUG-TPZ-CTR-CAZ-AZT-COT-CHL-LEV | 6 | 1 (4.16) | |
| AUG-TPZ-CTX-CTR-CAZ-GEN-NET-COT-LEV | 5 | 9 | 1 (4.16) |
| AUG-TPZ-CTX-CTR-CAZ-AZT-GEN-CHL-LEV | 6 | 10 | 2 (8.33) |
| AUG-TPZ-CTX-CTR-CAZ-AZT-GEN-NET-COT | 6 | 1 (4.16) | |
| AUG-TPZ-CTX-CTR-CAZ-AZT-GEN-NET-COT-LEV | 6 | 11 | 5 (20.83) |
| AUG-TPZ-CTR-CAZ-AZT-GEN-NET-COT-CHL-LEV | 7 | 1 (4.16) | |
| AUG-TPZ-CTX-CTR-CAZ-AZT-GEN-NET-COT-CHL-LEV | 7 | 12 | 1 (4.16) |
| Bacterial Species | N (%) | Resistance Mechanism | |||||
|---|---|---|---|---|---|---|---|
| ESBL | ESBL+AmpC | ESBL + Loss of Porin | AmpC | OXA-48 | KPC | ||
| E. coli | 14 | 5 (35.71) | 1 (6.67) | 1 (6.67) | 2 (13.33) | 3 (20.0) | 1 (6.67) |
| K. pneumoniae | 5 | 4 (80.0) | 3 (60.0) | 0 (0) | 3 (60.0) | 4 (80.0) | 0 (0) |
| E. gergoviae | 1 | 1 (100) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| E. aerogenes | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| S. fonticola | 1 | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 1 (100) | 0 (0) |
| K. oxytoca | 1 | 1 (100) | 0 (0) | 1 (100) | 0 (0) | 1 (100) | 0 (0) |
| R. ornithynolitica | 1 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Overall n (%) | 25 | 12 (48.0) | 6 (24.0) | 2 (8.0) | 5 (20.0) | 11 (44.0) | 1 (4.0) |
| MDR-E Species | MDR+ESBL Positive n (%) | β-Lactamases Genes | ||
|---|---|---|---|---|
| blaCTX-M n (%) | blaCTX-M-group-1,2,9 n (%) | blaTEM n (%) | ||
| Overall, n = 25 | 12 (48.0) | 12 (48.0) | 12 (48.0) | 18 (72.0) |
| E. coli; (n = 14) | 5 (35.71) | 6(42.85) | 6(42.85) | 11 (78.57) |
| K. pneumoniae; (n = 5) | 4 (80.0) | 4 (80.0) | 4 (80.0) | 4 (80.0) |
| K. oxytoca; (n = 2) | 1 (50.0) | 1 (50.0) | 1 (50.0) | 1 (50.0) |
| E. aerogenes; (n = 1) | 0 (0) | 0 (0) | 0 (0) | 1 (100) |
| E. gergoviae; (n = 1) | 1 (100) | 0 (0) | 0 (0) | 1 (100) |
| S. fonticola; (n = 1) | 1 (100) | 1 (100) | 1 (100) | 1 (100) |
| MDR-E Species (n) | Phenotypic Screennig of ESBL-PE | bla Resistances Genes | N (%) |
|---|---|---|---|
| E. coli (n = 14) | Positive | blaCTX-M, blaCTX-M-group-1,2,9, blaTEM | 4 (28.57%) |
| blaCTX-M, blaCTX-M-group-1,2,9 | 1 (7.14%) | ||
| Negative | blaTEM | 6 (42.8%) | |
| Negative | blaCTX-M, blaCTX-M-group-1,2,9, blaTEM | 1 (7.14%) | |
| K. pneumoniae (n = 5) | Positive | blaCTX-M, blaCTX-M-group-1,2,9, blaTEM | 4 (80.0%) |
| K. oxytoca (n = 2) | Positive | blaCTX-M, blaCTX-M-group-1,2,9 | 1 (50.0%) |
| E. aerogenes (n = 1) | Negative | blaTEM | 1 (100%) |
| E. gergoviae (n = 1) | Positive | blaTEM | 1 (100%) |
| S. fonticola (n = 1) | Positive | blaCTX-M, blaCTX-M-group-1,2,9, blaTEM | 1 (100%) |
| Bla Targeted | Primer Name | Sequence (5′-3′) | Amplicon Size (bp) | Annealing Temperatures | References |
|---|---|---|---|---|---|
| TEM | TEMF TEMR | CATTTCCGTGTCGCCCTTATTC CCAATGCTTAATCAGTGAGGC | 846 pb | 49.2 °C | [22] (p. 712) |
| CTX-M | CTX-MU1 CTX-MU2 | CGATGTGCAGTACCAGTAA TTAGTGACCAGAATCAGCGG | 585 pb | 46.6 °C | [23] (p. 1319) |
| CTX-M-group-1,2,9 | CTX-MA1 CTX-MA2 | SCSATGTGCAGYACCAGTAA CCGCRATATGKTTGGTGGTG | 585 pb | 56.2 °C | [24] (p. 29) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zemtsa, R.J.; Noubom, M.; Founou, L.L.; Dimani, B.D.; Koudoum, P.L.; Mbossi, A.D.; Kouanfack, C.; Founou, R.C. Multidrug-Resistant and Extended-Spectrum β-Lactamase (ESBL) - Producing Enterobacterales Isolated from Carriage Samples among HIV Infected Women in Yaoundé, Cameroon. Pathogens 2022, 11, 504. https://doi.org/10.3390/pathogens11050504
Zemtsa RJ, Noubom M, Founou LL, Dimani BD, Koudoum PL, Mbossi AD, Kouanfack C, Founou RC. Multidrug-Resistant and Extended-Spectrum β-Lactamase (ESBL) - Producing Enterobacterales Isolated from Carriage Samples among HIV Infected Women in Yaoundé, Cameroon. Pathogens. 2022; 11(5):504. https://doi.org/10.3390/pathogens11050504
Chicago/Turabian StyleZemtsa, Ravalona Jessica, Michel Noubom, Luria Leslie Founou, Brice Davy Dimani, Patrice Landry Koudoum, Aurelia Djeumako Mbossi, Charles Kouanfack, and Raspail Carrel Founou. 2022. "Multidrug-Resistant and Extended-Spectrum β-Lactamase (ESBL) - Producing Enterobacterales Isolated from Carriage Samples among HIV Infected Women in Yaoundé, Cameroon" Pathogens 11, no. 5: 504. https://doi.org/10.3390/pathogens11050504
APA StyleZemtsa, R. J., Noubom, M., Founou, L. L., Dimani, B. D., Koudoum, P. L., Mbossi, A. D., Kouanfack, C., & Founou, R. C. (2022). Multidrug-Resistant and Extended-Spectrum β-Lactamase (ESBL) - Producing Enterobacterales Isolated from Carriage Samples among HIV Infected Women in Yaoundé, Cameroon. Pathogens, 11(5), 504. https://doi.org/10.3390/pathogens11050504







