Establishment and Application of a Multiplex PCR Assay for the Rapid Detection of Rhizoctonia solani Anastomosis Group (AG)-3PT, the Pathogen Causing Potato Black Scurf and Stem Canker
Abstract
1. Introduction
2. Results
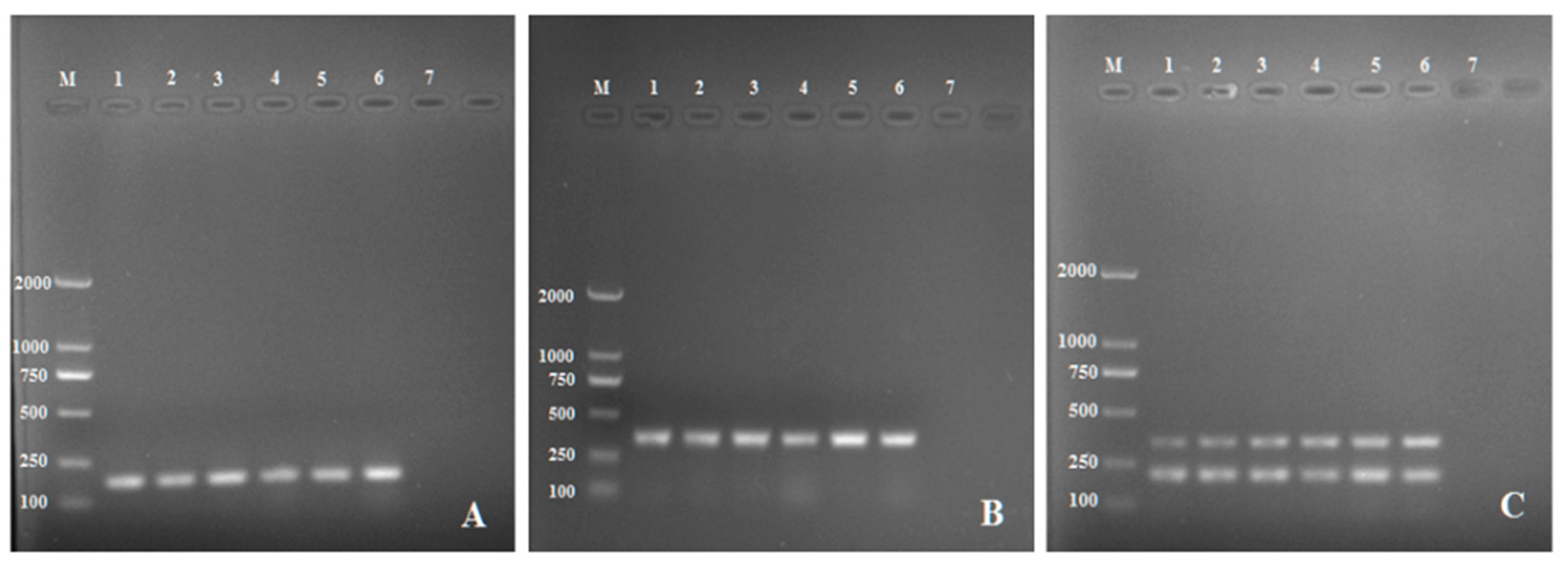
2.1. Detection of R. solani AG-3 in Single PCR and Multiplex PCR
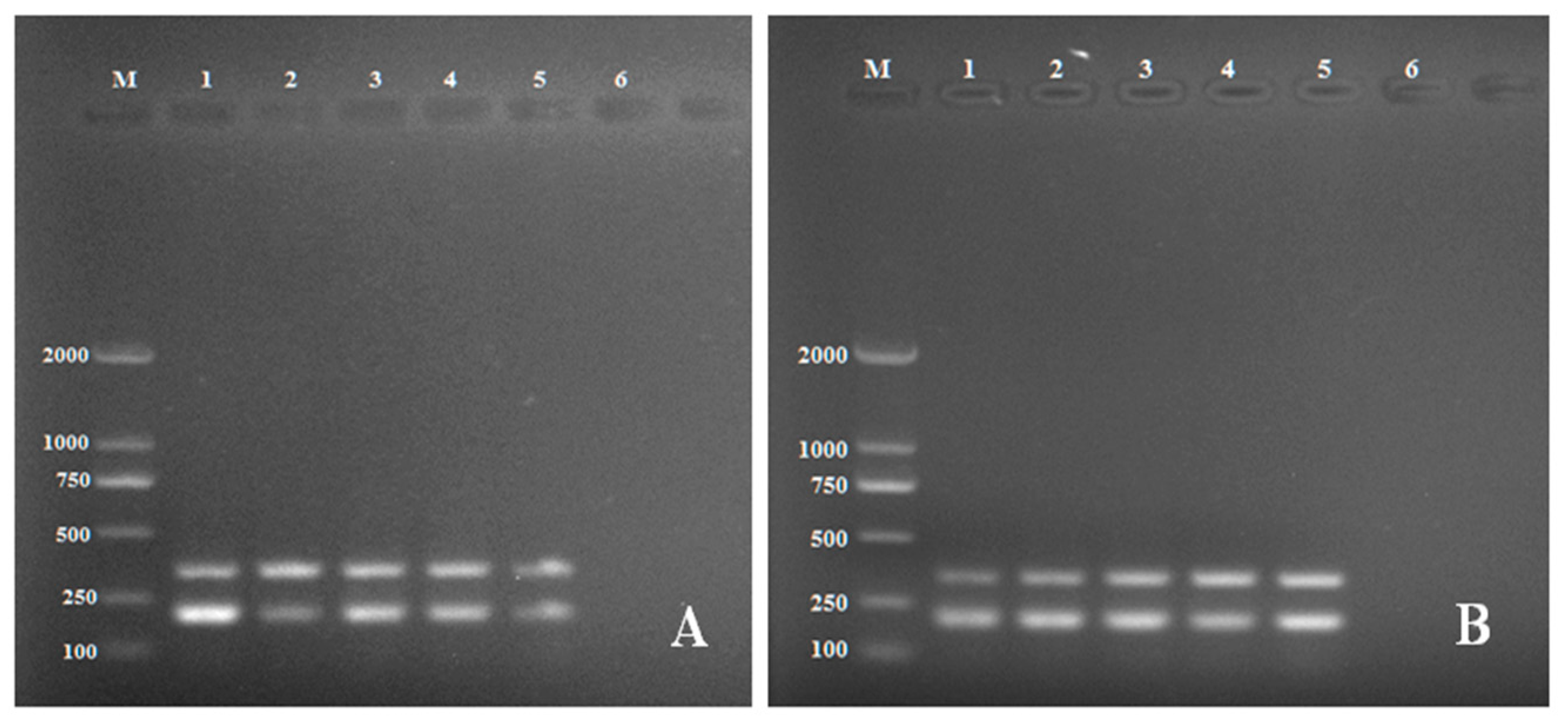
2.2. Verification of Primer Specificity by Multiplex PCR
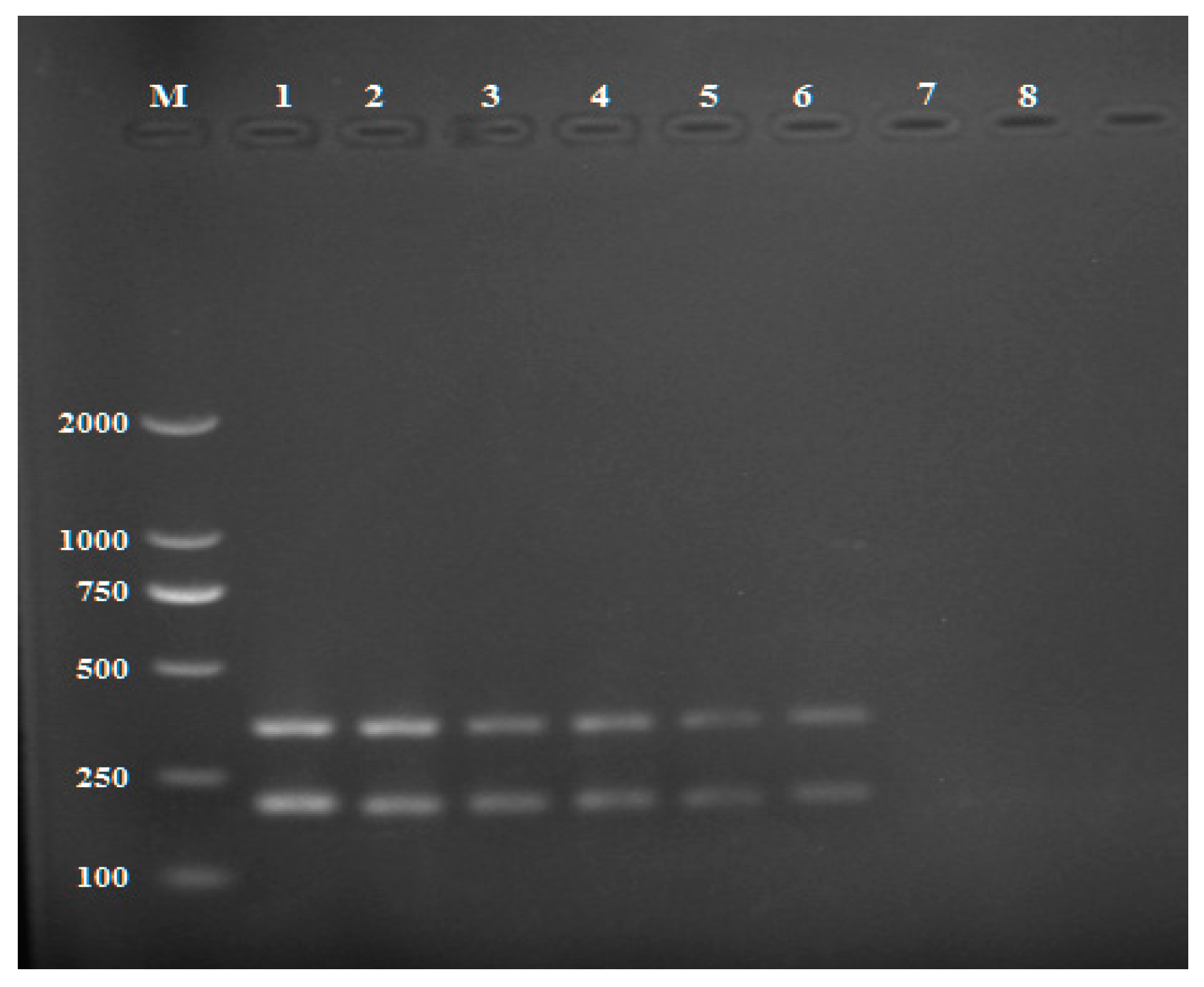
2.3. Detection of Rhizoctonia solani in Artificially Infected Soils and Tubers by Multiplex PCR
3. Discussion
4. Materials and Methods
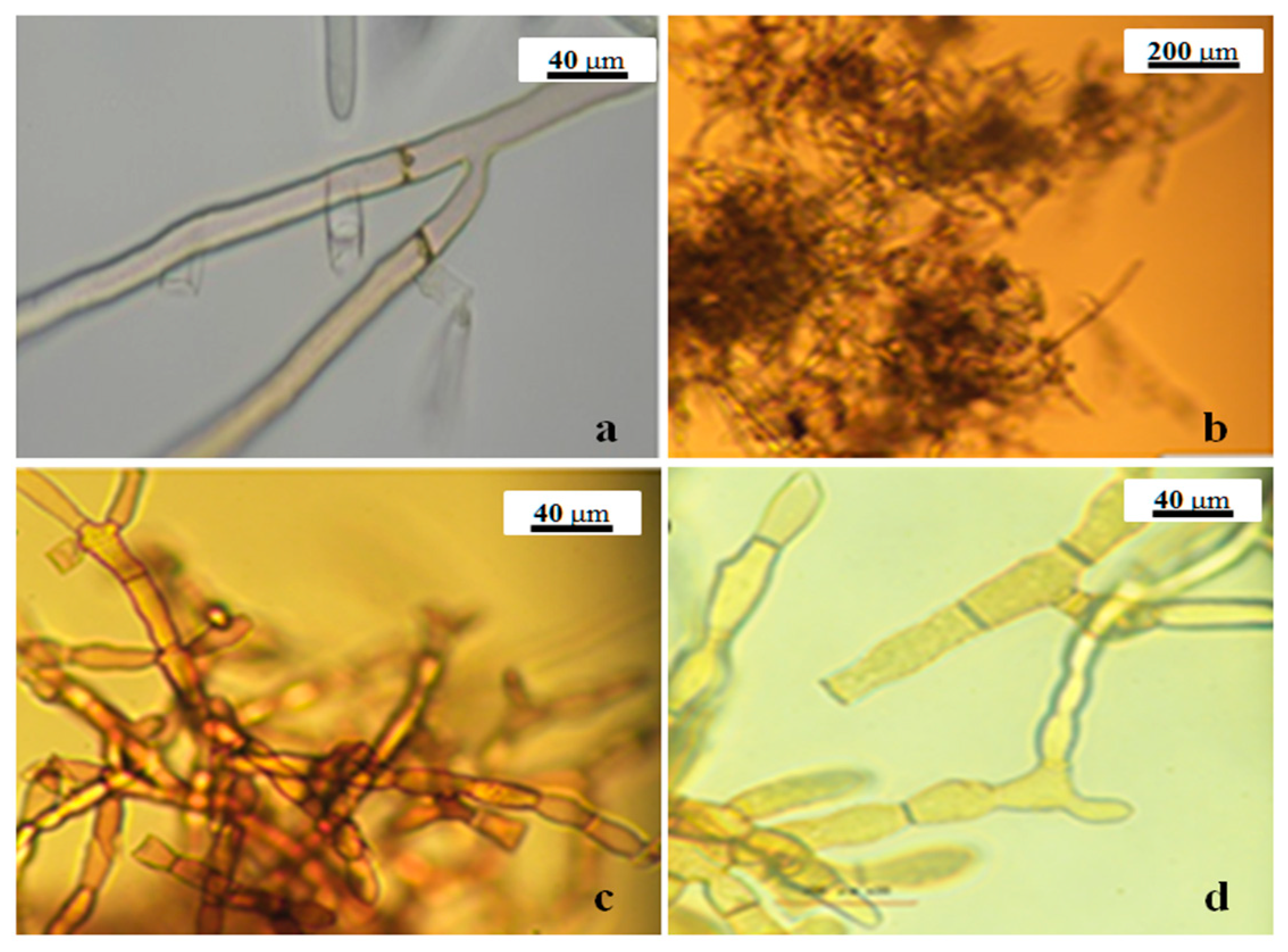
4.1. Source of Isolates and Morphological Identification
4.2. DNA Extraction
4.3. Primer Design
4.4. Detection of Rhizoctonia Solani by Single PCR
4.5. Multiplex PCR Detection of R. solani in Artificially Infected Soil
4.6. Multiplex PCR Detection of R. solani in Infected Tubers
4.7. Optimization of Multiplex PCR
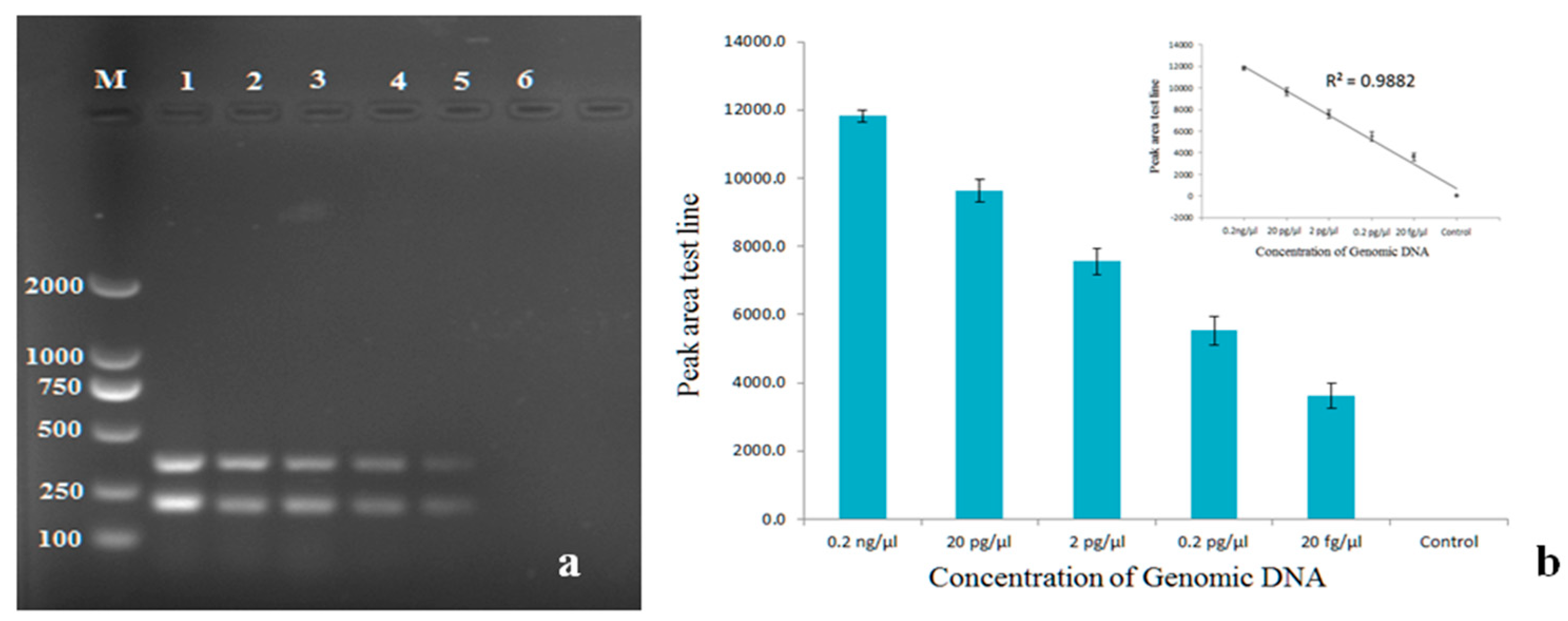
4.8. Sensitivity Assay
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xia, Y.; Fei, B.; He, J.; Zhou, M.; Zhang, D.; Pan, L.; Li, S.; Liang, Y.; Wang, L.; Zhu, J.; et al. Transcriptome analysis reveals the host selection fitness mechanisms of the Rhizoctonia solani AG1IA pathogen. Sci. Rep. 2017, 7, 10120. [Google Scholar] [CrossRef]
- Errampalli, D.; Johnston, H.W. Control of tuber-borne black scurf [Rhizoctonia solani] and common scab [Streptomyces scabies] of potatoes with a combination of sodium hypochlorite and thiophanate-methyl preplanting seed tuber treatment. Can. J. Plant Pathol. 2001, 23, 68–77. [Google Scholar] [CrossRef]
- Ajayi-Oyetunde, O.O.; Bradley, C.A. Rhizoctonia solani: Taxonomy, population biology and management of Rhizoctonia seedling disease of soybean. Plant Pathol. 2017, 67, 3–17. [Google Scholar] [CrossRef]
- Ogoshi, A. Ecology and Pathogenicity of Anastomosis and Intraspecific Groups of Rhizoctonia solani Kuhn. Annu. Rev. Phytopathol. 1987, 25, 125–143. [Google Scholar] [CrossRef]
- Tsror, L. Biology, Epidemiology and Management of Rhizoctonia solani on Potato. J. Phytopathol. 2010, 158, 649–658. [Google Scholar] [CrossRef]
- Tuncer, S.; Eken, C. Anastomosis grouping of Rhizoctonia solani and binucleate Rhizoctonia spp. isolated from pepper in Erzincan, Turkey. Plant Prot. Sci. 2013, 49, 127–131. [Google Scholar] [CrossRef]
- Taheri, P.; Tarighi, S. The Role of Pathogenesis-Related Proteins in the Tomato-Rhizoctonia solani Interaction. J. Bot. 2012, 2012, 137037. [Google Scholar] [CrossRef]
- Carling, D.; Baird, R.; Gitaitis, R.; Brainard, K.; Kuninaga, S. Characterization of AG-13, a newly reported anastomosis group of Rhizoctonia solani. Phytopathology 2002, 92, 893–899. [Google Scholar] [CrossRef]
- Sneh, B.; Jabaji-Hare, S.; Neate, S.; Dijst, G. Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Prabha, S.; Vignesh, A.; Murugesan, K. Biological control of damping off and stem rot of tomato (Lycopersicon esculentum Mill.) using an antagonistic actinomycete, Saccharopolyspora sp. Int. J. Agric. Sci. Res. 2014, 4, 55–65. [Google Scholar]
- Ferrucho, R.L.; Cifuentes, J.M.; Ceresini, P.; García-Domínguez, C. Rhizoctonia solani AG-3PT is the major pathogen associated with potato stem canker and black scurf in Colombia. Agron. Colomb. 2012, 30, 204–213. [Google Scholar]
- Das, S.; Shah, F.A.; Butler, R.C.; Falloon, R.E.; Stewart, A.; Raikar, S.; Pitman, A.R. Genetic variability and pathogenicity of R hizoctonia solani associated with black scurf of potato in New Zealand. Plant Pathol. 2014, 63, 651–666. [Google Scholar] [CrossRef]
- Djébali, N.; Belhassen, T. Field study of the relative susceptibility of eleven potato (Solanum tuberosum L.) varieties and the efficacy of two fungicides against Rhizoctonia solani attack. Crop Prot. 2010, 29, 998–1002. [Google Scholar] [CrossRef]
- El Bakali, A.M.; Martín, M.P. Black scurf of potato. Mycologist 2006, 20, 130–132. [Google Scholar] [CrossRef]
- Kankam, F.; Larbi-Koranteng, S.; Adomako, J. Rhizoctonia Disease of Potato: Epidemiology, Toxin Types and Management. Egypt. J. Phytopathol. 2021, 49, 197–209. [Google Scholar] [CrossRef]
- Banville, G.J. Yield losses and damage to potato plants caused by Rhizoctonia solani Kuhn. Am. Potato J. 1989, 66, 821–834. [Google Scholar] [CrossRef]
- Kiptoo, J.; Abbas, A.; Bhatti, A.M.; Usman, H.M.; Shad, M.A.; Umer, M.; Atiq, M.N.; Alam, S.M.; Ateeq, M.; Khan, M.; et al. Rhizoctonia solani of Potato and Its Management: A review. Plant Prot. 2021, 5, 157–169. [Google Scholar] [CrossRef]
- Budge, G.; Shaw, M.; Colyer, A.; Pietravalle, S.; Boonham, N. Molecular tools to investigate Rhizoctonia solani distribution in soil. Plant Pathol. 2009, 58, 1071–1080. [Google Scholar] [CrossRef]
- Hariharan, G.; Prasannath, K. Recent Advances in Molecular Diagnostics of Fungal Plant Pathogens: A Mini Review. Front. Cell. Infect. Microbiol. 2021, 10, 829. [Google Scholar] [CrossRef]
- Singh, Y.; Singh, J.; Pandey, A. Molecular markers in diagnosis and management of fungal pathogens: A review. Int. J. Adv. Biotechnol. Res. 2013, 4, 180–188. [Google Scholar]
- Nikitin, M.; Deych, K.; Grevtseva, I.; Girsova, N.; Kuznetsova, M.; Pridannikov, M.; Dzhavakhiya, V.; Statsyuk, N.; Golikov, A. Preserved Microarrays for Simultaneous Detection and Identification of Six Fungal Potato Pathogens with the Use of Real-Time PCR in Matrix Format. Biosensors 2018, 8, 129. [Google Scholar] [CrossRef]
- Mancini, V.; Murolo, S.; Romanazzi, G. Diagnostic methods for detecting fungal pathogens on vegetable seeds. Plant Pathol. 2016, 65, 691–703. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of Apple Leaf Diseases Based on Deep Convolutional Neural Networks. Symmetry 2017, 10, 11. [Google Scholar] [CrossRef]
- Häusera, F.; Gökceb, S.; Wernera, G.; Danckwardta, S.; Sollfranka, S.; Neukircha, C.; Beyerc, V.; Hennermannb, J.B.; Lacknera, K.J.; Mengelb, E.; et al. A non-invasive diagnostic assay for rapid detection and characterization of aberrant mRNA-splicing by nonsense mediated decay inhibition. Mol. Genet. Metab. 2020, 130, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.K.; Ballard, G.; Stahl, D.A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 1993, 59, 1354–1360. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, C.; Fung, C.; Li, X.-F. Quantification of Viable but Nonculturable Escherichia coli O157:H7 by Targeting the rpoS mRNA. Anal. Chem. 2010, 82, 2612–2615. [Google Scholar] [CrossRef]
- Lievens, B.; Thomma, B.P.H.J. Recent Developments in Pathogen Detection Arrays: Implications for Fungal Plant Pathogens and Use in Practice. Phytopathology 2005, 95, 1374–1380. [Google Scholar] [CrossRef]
- Walcott, R.R. Detection of Seedborne Pathogens. HortTechnology 2003, 13, 40–47. [Google Scholar] [CrossRef]
- Ward, E.; Foster, S.J.; Fraaije, B.A.; Mccartney, H.A. Plant pathogen diagnostics: Immunological and nucleic acid-based approaches. Ann. Appl. Biol. 2004, 145, 1–16. [Google Scholar] [CrossRef]
- Chilvers, M.I. Molecular Diagnostics in Plant Disease Diagnostic Clinics… What’s the Status? Fungal Genom. Biol. 2012, 2, e102. [Google Scholar] [CrossRef]
- Kwak, H.-R.; Kim, M.-K.; Shin, J.-C.; Lee, Y.-J.; Seo, J.-K.; Lee, H.-U.; Jung, M.-N.; Kim, S.-H.; Choi, H.-S. The Current Incidence of Viral Disease in Korean Sweet Potatoes and Development of Multiplex RT-PCR Assays for Simultaneous Detection of Eight Sweet Potato Viruses. Plant Pathol. J. 2014, 30, 416–424. [Google Scholar] [CrossRef]
- López, M.M.; Bertolini, E.; Olmos, A.; Caruso, P.; Gorris, M.T.; Llop, P.; Penyalver, R.; Cambra, M. Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 2003, 6, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Makkouk, K.; Kumari, S. Molecular diagnosis of plant viruses. Arab. J. Plant Prot. 2006, 24, 135–138. [Google Scholar]
- Lees, A.K.; Cullen, D.W.; Sullivan, L.; Nicolson, M.J. Development of conventional and quantitative real-time PCR assays for the detection and identification of Rhizoctonia solani AG-3 in potato and soil. Plant Pathol. 2002, 51, 293–302. [Google Scholar] [CrossRef]
- Capote, N.; Pastrana, A.M.; Aguado, A.; Sánchez-Torres, P. Molecular tools for detection of plant pathogenic fungi and fungicide resistance. In Plant Pathology; InTech: Rijeka, Croatia, 2012; pp. 151–202. [Google Scholar]
- Aslam, S.; Tahir, A.; Aslam, M.F.; Alam, M.W.; Shedayi, A.A.; Sadia, S. Recent advances in molecular techniques for the identification of phytopathogenic fungi–a mini review. J. Plant Interact. 2017, 12, 493–504. [Google Scholar] [CrossRef]
- Lee, S.B.; Taylor, J.W. Isolation of DNA from Fungal Mycelia and Single Spores. In PCR Protocols: A Guide to Methods and Applications; Academic Press: San Diego, CA, USA, 1990; pp. 282–287. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef]
- James, D. A simple and reliable protocol for the detection of apple stem grooving virus by RT–PCR and in a multiplex PCR assay. J. Virol. Methods 1999, 83, 1–9. [Google Scholar] [CrossRef]
- Williams, K.; Blake, S.; Sweeney, A.; Singer, J.T.; Nicholson, B.L. Multiplex Reverse Transcriptase PCR Assay for Simultaneous Detection of Three Fish Viruses. J. Clin. Microbiol. 1999, 37, 4139–4141. [Google Scholar] [CrossRef]
- Nakadomari, G.H.; Charalo, A.C.; Pavan, A.C.L.; Vignoto, V.K.C.; Sfaciotte, R.A.P.; Wosiacki, S.R. Multiplex-PCR for detection of β-lactam resistance in Staphylococcus spp. Revista de Ciência Veterinária E Saúde Pública 2019, 6, 262–275. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef]
- Brownie, J. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997, 25, 3235–3241. [Google Scholar] [CrossRef]
- Fang, Y.; Ramasamy, R.P. Current and Prospective Methods for Plant Disease Detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Bruehl, G.W. Plant Pathology, a Changing Profession in a Changing World. Annu. Rev. Phytopathol. 1991, 29, 1–13. [Google Scholar] [CrossRef]
- Hrytsev, O.; Shevchenko, J.; Vorobiova, N.; Skivka, L. Multiplex-touchdown pcr for rapid simultaneous detection of Rhizoctonia cerealis AND Rhizoctonia solani. Biotechnol. Acta 2019, 12, 75–81. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.; Zuo, J.; Gong, J.; Hu, J.; Jiang, W.; Mi, R.; Huang, Y.; Chen, Z.; Phouthapane, V.; Qi, K.; et al. Development of a multiplex PCR assay for the simultaneous and rapid detection of six pathogenic bacteria in poultry. AMB Express 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The its Region of Nuclear Ribosomal DNA: A Valuable Source of Evidence on Angiosperm Phylogeny. Ann. Mo. Bot. Gard. 1995, 82, 247. [Google Scholar] [CrossRef]
- Fitzpatrick, T.B.; Amrhein, N.; Kappes, B.; Macheroux, P.; Tews, I.; Raschle, T. Two independent routes of de novo vitamin B6 biosynthesis: Not that different after all. Biochem. J. 2007, 407, 1–13. [Google Scholar] [CrossRef]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: A practical approach. J. Clin. Lab. Anal. 2002, 16, 47–51. [Google Scholar] [CrossRef]



| Code | Host | Origin | Anastomosis Group/Species |
|---|---|---|---|
| AG-3 | Potato | Gansu Province | Rhizoctonia solani AG-3 |
| AG-1 | Rice | Yunnan Province | Rhizoctonia solani AG-1 |
| AG-4 | Cotton | Hubei Province | Rhizoctonia solani AG-4 |
| PSI | Glycine max | Fujian Province | Phytophthora sojae |
| PCI | Capsicum annuum | Fujian Province | Phytophthora capsici |
| PNI | Nicotiana tabacum | Chongqing Province | Phytophthora nicotianae |
| AA | Potato | Fujian Province | Alternaria alternata |
| AT | Potato | Fujian Province | Alternaria tenuissima |
| AS | Potato | Shandong Province | Alternaria solani |
| Target Gene | Primer Name | Sequence (5’-3’) | Amplicon Size (bp) |
|---|---|---|---|
| Endopolygalacturonase | RsE-F | GCACTATCTCGTCGTTGA | 151 |
| RsE-R | ACTCCGAACTTGACATCTC | ||
| Pyridoxine biosynthesis protein PDX1 | RsP-F | GCGTTTGCCAGTTGTCAG | 342 |
| RsP-R | CACATAGTCAGCCCAACCA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iradukunda, L.; Wang, Y.-P.; Nkurikiyimfura, O.; Wang, T.; Yang, L.-N.; Zhan, J. Establishment and Application of a Multiplex PCR Assay for the Rapid Detection of Rhizoctonia solani Anastomosis Group (AG)-3PT, the Pathogen Causing Potato Black Scurf and Stem Canker. Pathogens 2022, 11, 627. https://doi.org/10.3390/pathogens11060627
Iradukunda L, Wang Y-P, Nkurikiyimfura O, Wang T, Yang L-N, Zhan J. Establishment and Application of a Multiplex PCR Assay for the Rapid Detection of Rhizoctonia solani Anastomosis Group (AG)-3PT, the Pathogen Causing Potato Black Scurf and Stem Canker. Pathogens. 2022; 11(6):627. https://doi.org/10.3390/pathogens11060627
Chicago/Turabian StyleIradukunda, Linda, Yan-Ping Wang, Oswald Nkurikiyimfura, Tian Wang, Li-Na Yang, and Jiasui Zhan. 2022. "Establishment and Application of a Multiplex PCR Assay for the Rapid Detection of Rhizoctonia solani Anastomosis Group (AG)-3PT, the Pathogen Causing Potato Black Scurf and Stem Canker" Pathogens 11, no. 6: 627. https://doi.org/10.3390/pathogens11060627
APA StyleIradukunda, L., Wang, Y.-P., Nkurikiyimfura, O., Wang, T., Yang, L.-N., & Zhan, J. (2022). Establishment and Application of a Multiplex PCR Assay for the Rapid Detection of Rhizoctonia solani Anastomosis Group (AG)-3PT, the Pathogen Causing Potato Black Scurf and Stem Canker. Pathogens, 11(6), 627. https://doi.org/10.3390/pathogens11060627






