A Whole Genome Sequencing-Based Epidemiological Investigation of a Pregnancy-Related Invasive Listeriosis Case in Central Italy
Abstract
:1. Introduction
2. Results
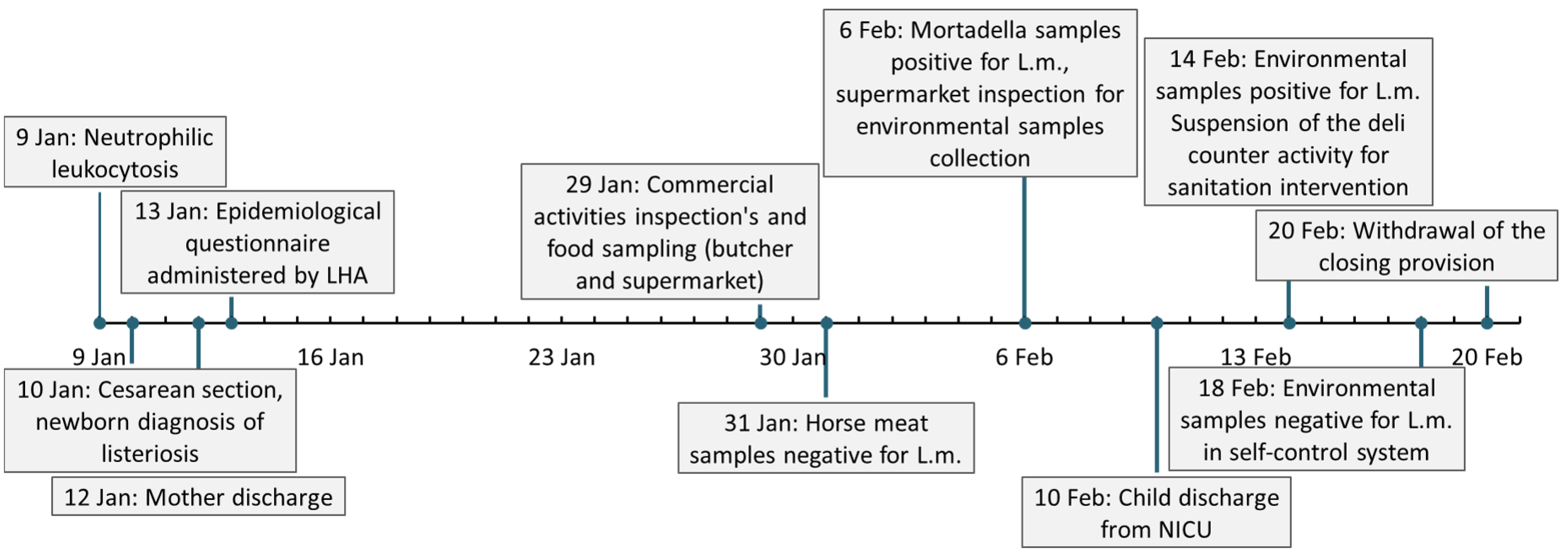
2.1. Case Presentation
2.2. Retrospective Epidemiological Investigation
2.3. Bacterial Identification and Serotyping
2.4. In Silico Analysis
3. Discussion
4. Materials and Methods
4.1. Food and Environmental Sampling
4.2. Microbiological Methods for Bacterial Identification and Serotyping
4.3. Whole Genome Sequencing and In Silico Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kathariou, S. Listeria monocytogenes Virulence and Pathogenicity, a Food Safety Perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Lorber, B. Listeria monocytogenes. In Principles and Practice of Infectious Diseases, 5th ed.; Mandell, G.L., Bennett, J.E., Dolin, R., Eds.; Churchill Livingstone: London, UK, 2000. [Google Scholar]
- Schlech, W.F.; Lavigne, P.M.; Bortolussi, R.A.; Allen, A.C.; Haldane, E.V.; Wort, A.J.; Hightower, A.W.; Johnson, S.E.; King, S.H.; Nicholls, E.S.; et al. Epidemic Listeriosis—Evidence for Transmission by Food. N. Engl. J. Med. 1983, 308, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Hafner, L.; Pichon, M.; Burucoa, C.; Nusser, S.H.A.; Moura, A.; Garcia-Garcera, M.; Lecuit, M. Listeria monocytogenes faecal carriage is common and depends on the gut microbiota. Nat. Commun. 2021, 12, 6826. [Google Scholar] [CrossRef]
- Buffie, C.G.; Pamer, E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013, 13, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Doganay, M. Listeriosis: Clinical presentation. FEMS Immunol. Med. Microbiol. 2003, 35, 173–175. [Google Scholar] [CrossRef]
- Matle, I.; Mbatha, K.R.; Madoroba, E. A review of Listeria monocytogenes from meat and meat products: Epidemiology, virulence factors, antimicrobial resistance and diagnosis. Onderstepoort J. Vet. Res. 2020, 87, 1–20. [Google Scholar] [CrossRef]
- Desai, A.N.; Anyoha, A.; Madoff, L.C.; Lassmann, B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: A review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Stout, A.; Van Stelten-Carlson, A.; Marquis, H.; Ballou, M.; Reilly, B.; Loneragan, G.H.; Nightingale, K.; Ivanek, R. Public health impact of foodborne exposure to naturally occurring virulence-attenuated Listeria monocytogenes: Inference from mouse and mathematical models. Interface Focus 2020, 10, 20190046. [Google Scholar] [CrossRef] [Green Version]
- Russini, V.; Spaziante, M.; Zottola, T.; Fermani, A.G.; Di Giampietro, G.; Blanco, G.; Fabietti, P.; Marrone, R.; Parisella, R.; Parrocchia, S.; et al. A Nosocomial Outbreak of Invasive Listeriosis in An Italian Hospital: Epidemiological and Genomic Features. Pathogens 2021, 10, 591. [Google Scholar] [CrossRef] [PubMed]
- Tortajada, C.; Porta, R.; Riba, M.; Santoma, M.J.; Palacín, E.; Español, M. Nosocomial outbreak due to Listeria monocytogenes in a neonatal unit. Enferm. Infecc. Microbiol. Clin. 2012, 30, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Schuchat, A.; Lizano, C.; Broome, C.V.; Swaminathan, B.; Kim, C.; Winn, K. Outbreak of neonatal listeriosis associated with mineral oil. Pediatr. Infect. Dis. J. 1991, 10, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Elsner, H.-A.; Kaulfers, P.-M.; Tenschert, W.; Fischer, L. Nosocomial infections by Listeria monocytogenes: Analysis of a cluster of septicemias in immunocompromised patients. Infection 1997, 25, 135–139. [Google Scholar] [CrossRef]
- Voetsch, A.C.; Angulo, F.J.; Jones, T.F.; Moore, M.R.; Madon, C.; McCarthy, P.; Shiferaw, B.; Megginson, M.B.; Hurd, S.; Anderson, B.J.; et al. Reduction in the incidence of invasive listeriosis in foodborne diseases active surveillance network sites, 1996–2003. Clin. Infect. Dis. 2007, 44, 513–520. [Google Scholar] [CrossRef] [Green Version]
- Janakiraman, V. Listeriosis in pregnancy: Diagnosis, treatment, and prevention. Rev. Obstet. Gynecol. 2008, 1, 179–185. [Google Scholar]
- Jackson, K.A.; Iwamoto, M.; Swerdlow, D. Pregnancy-associated listeriosis. Epidemiol. Infect. 2010, 138, 1503–1509. [Google Scholar] [CrossRef] [Green Version]
- Charlier, C.; Disson, O.; Lecuit, M. Maternal-neonatal listeriosis. Virulence 2020, 11, 391–397. [Google Scholar] [CrossRef]
- Mateus, T.; Silva, J.; Maia, R.L.; Teixeira, P.; Canellada, A.; Malek, A.; Peterson, C.M. Listeriosis during Pregnancy: A Public Health Concern. Obstet. Gynecol. 2013, 2013, 851712. [Google Scholar] [CrossRef] [Green Version]
- Smith, J.L. Foodborne Infections during Pregnancy. J. Food Prot. 1999, 62, 818–829. [Google Scholar] [CrossRef]
- Ogunmodede, F.; Jones, J.L.; Scheftel, J.; Kirkland, E.; Schulkin, J.; Lynfield, R. Listeriosis Prevention Knowledge Among Pregnant Women in the USA. Infect. Dis. Obstet. Gynecol. 2005, 13, 11–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leber, A.; Zenclussen, M.L.; Teles, A.; Brachwitz, N.; Casalis, P.; El-Mousleh, T.; Jensen, F.; Woidacki, K.; Zenclussen, A.C. Pregnancy: Tolerance and Suppression of Immune Responses. Methods Mol. Biol. 2011, 677, 397–417. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, H. Listeria infection in women. Prim. Care Update Ob. Gyns. 2000, 7, 40–45. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Vital signs: Listeria illnesses, deaths, and outbreaks—United States, 2009–2011. Morb. Mortal. Wkly. Rep. 2013, 62, 448–452. [Google Scholar]
- Lamont, R.F.; Sobel, J.; Mazaki-Tovi, S.; Kusanovic, J.P.; Vaisbuch, E.; Kim, S.K.; Uldbjerg, N.; Romero, R. Listeriosis in human pregnancy: A systematic review. J. Perinat. Med. 2011, 39, 1–7. [Google Scholar] [CrossRef]
- McLauchlin, J. Human listeriosis in Britain, 1967–85, a summary of 722 cases: 2. Listeriosis in non-pregnant individuals, a changing pattern of infection and seasonal incidence. Epidemiol. Infect. 1990, 104, 191–201. [Google Scholar] [CrossRef]
- Serventi, L.; Curi, B.; Johns, R.; Silva, J.; Bainbridge, R.; Gaither, K. Pregnancy Complicated by Listeria Monocytogenes: A Case Report and Review of the Literature. J. Natl. Med. Assoc. 2020, 112, 428–432. [Google Scholar] [CrossRef]
- De Luca, C.; Donati, L.; D’Oria, L.; Licameli, A.; Pellegrino, M.; De Santis, M. Listeria Infection in Pregnancy: A Review of Literature. Open Infect. Dis. J. 2015, 9, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Mylonakis, E.; Paliou, M.; Hohmann, E.L.; Calderwood, S.B.; Wing, E.J. Listeriosis during pregnancy: A case series and review of 222 cases. Medicine 2002, 81, 260–269. [Google Scholar] [CrossRef]
- Reiss, H.J.; Krebs, A. Septic granulomatosis in infants, a fetal sepsis caused by a specific pathogen. Klin. Wochenschr. 1951, 29, 29. [Google Scholar] [CrossRef]
- Johnston, W.H.; Morton, S.A.; Wong, M.H.; Roy, T.E. Septicaemia of the newborn due to listeria monocytogenes. Can. Med. Assoc. J. 1955, 73, 402–405. [Google Scholar] [CrossRef] [PubMed]
- Becroft, D.M.O.; Farmer, K.; Seddon, R.J.; Sowden, R.; Stewart, J.H.; Vines, A.; Wattie, D.A. Epidemic Listeriosis in the Newborn. Br. Med. J. 1971, 3, 747–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLauchlin, J. Human listeriosis in Britain, 1967–85, a summary of 722 cases: 1. Listeriosis during pregnancy and in the newborn. Epidemiol. Infect. 1990, 104, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Galetta, P.; Dionisi, A.M.; Filetici, E.; Benedetti, I.; Arena, S.; Owczarek, S.; Lana, S.; Bella, A.; Scavia, G.; Rizzo, C.; et al. Enter-net: Sorveglianza delle infezioni da patogeni enterici. Not Ist Super Sanità 2008, 21, 11–17. [Google Scholar]
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5926. [Google Scholar] [CrossRef] [Green Version]
- Filipello, V.; Amato, E.; Gori, M.; Huedo, P.; Ciceri, G.; Lomonaco, S.; Pontello, M. Epidemiology and Molecular Typing of Pregnancy-Associated Listeriosis Cases in Lombardy, Italy, over a 10-Year Period (2005–2014). Infect. Dis. Obstet. Gynecol. 2017, 2017, 6479121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yde, M.; Botteldoorn, N.; Bertrand, S.; Collard, J.M.; Dierick, K. Microbiological and molecular investigation of an increase of human listeriosis in Belgium, 2006–2007. Eurosurveillance 2010, 15, 5. [Google Scholar] [CrossRef]
- Mook, P.; Grant, K.A.; Little, C.L.; Kafatos, G.; Gillespie, I.A. Emergence of pregnancy-related listeriosis amongst ethnic minorities in England and Wales. Eurosurveillance 2010, 15, 17–23. [Google Scholar] [CrossRef]
- Girard, D.; Leclercq, A.; Laurent, E.; Lecuit, M.; de Valk, H.; Goulet, V. Pregnancy-related listeriosis in France, 1984 to 2011, with a focus on 606 cases from 1999 to 2011. Eurosurveillance 2014, 19, 20909. [Google Scholar] [CrossRef] [Green Version]
- Elinav, H.; Hershko-Klement, A.; Valinsky, L.; Jaffe, J.; Wiseman, A.; Shimon, H.; Braun, E.; Paitan, Y.; Block, C.; Sorek, R.; et al. Pregnancy-Associated Listeriosis: Clinical Characteristics and Geospatial Analysis of a 10-Year Period in Israel. Clin. Infect. Dis. 2014, 59, 953–961. [Google Scholar] [CrossRef] [Green Version]
- Herrador, Z.; Gherasim, A.; López-Vélez, R.; Benito, A. Listeriosis in Spain based on hospitalisation records, 1997 to 2015: Need for greater awareness. Eurosurveillance 2019, 24, 1800271. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Kasper, D.L.; Longo, D.L.; Braunwald, E.; Hauser, S.L.; Jameson, J.L.; Loscalzo, J. Harrison’s Principles of Internal Medicine, 17th ed.; McGraw-Hill Education: New York, NY, USA, 2008. [Google Scholar]
- Gaul, L.K.; Farag, N.H.; Shim, T.; Kingsley, M.A.; Silk, B.J.; Hyytia-Trees, E. Hospital-Acquired Listeriosis Outbreak Caused by Contaminated Diced Celery—Texas, 2010. Clin. Infect. Dis. 2013, 56, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Chen, X.; Payne, M.; Cao, X.; Wang, Y.; Zhang, J.; Deng, J.; Wang, H.; Zhang, Z.; Li, Q.; et al. Case report: Whole genome sequencing based investigation of maternal-neonatal listeriosis in Sichuan, China. BMC Infect. Dis. 2019, 19, 893. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Tourdjman, M.; Leclercq, A.; Hamelin, E.; Laurent, E.; Fredriksen, N.; van Cauteren, D.; Bracq-Dieye, H.; Thouvenot, P.; Vales, G.; et al. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg. Infect. Dis. 2017, 23, 1462–1470. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Knijn, A.; Michelacci, V.; Orsini, M.; Morabito, S. Advanced research infrastructure for experimentation in genomicS (ARIES): A lustrum of galaxy experience. bioRxiv 2020. [Google Scholar] [CrossRef]
- The Integrated Rapid Infectious Disease Analysis (IRIDA). Advanced Research Infrastructure for Experimentation in GenomicS (ARIES) Platform. Available online: https://irida.iss.it (accessed on 11 February 2022).
- Goulet, V.; King, L.A.; Vaillant, V.; de Valk, H. What is the incubation period for listeriosis? BMC Infect. Dis. 2013, 13, 11. [Google Scholar] [CrossRef] [Green Version]
- Harrand, A.S.; Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H. Evolution of listeria monocytogenes in a food processing plant involves limited single-nucleotide substitutions but considerable diversification by gain and loss of prophages. Appl. Environ. Microbiol. 2020, 86, e02493-19. [Google Scholar] [CrossRef]
- Ryan, E.M.; Gahan, C.G.M.; Hill, C. A significant role for Sigma B in the detergent stress response of Listeria monocytogenes. Lett. Appl. Microbiol. 2008, 46, 148–154. [Google Scholar] [CrossRef]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to Stress Conditions Encountered in Food and food processing environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef] [Green Version]
- Duchêne, S.; Holt, K.E.; Weill, F.X.; Le Hello, S.; Hawkey, J.; Edwards, D.J.; Fourment, M.; Holmes, E.C. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2016, 2, e000094. [Google Scholar] [CrossRef] [PubMed]
- Balmer, O.; Tanner, M. Prevalence and implications of multiple-strain infections. Lancet Infect. Dis. 2011, 11, 868–878. [Google Scholar] [CrossRef]
- Louhi, K.R.; Karvonen, A.; Rellstab, C.; Louhi, R.; Jokela, J. Prevalence of infection as a predictor of multiple genotype infection frequency in parasites with multiple-host life cycle. J. Anim. Ecol. 2013, 82, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.; Van Doorn, L.-J.; Nogueira, C.; Soares, J.M.; Pinho, C.; Figueira, P.; Quint, W.G.V.; Carneiro, F. Helicobacter pylori Genotypes Are Associated with Clinical Outcome in Portuguese Patients and Show a High Prevalence of Infections with Multiple Strains. Scand. J. Gastroenterol. 2001, 36, 128–135. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Monnier, A.L.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [Green Version]
- Muhterem-Uyar, M.; Ciolacu, L.; Wagner, K.-H.; Wagner, M.; Schmitz-Esser, S.; Stessl, B. New Aspects on Listeria monocytogenes ST5-ECVI Predominance in a Heavily Contaminated Cheese Processing Environment. Front. Microbiol. 2018, 9, 64. [Google Scholar] [CrossRef] [Green Version]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated From Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Chen, Y.; Luo, Y.; Curry, P.; Timme, R.; Melka, D.; Doyle, M.; Parish, M.; Hammack, T.S.; Allard, M.W.; Brown, E.W.; et al. Assessing the genome level diversity of Listeria monocytogenes from contaminated ice cream and environmental samples linked to a listeriosis outbreak in the United States. PLoS ONE 2017, 12, e0171389. [Google Scholar] [CrossRef]
- Du, X.j.; Zhang, X.; Wang, X.y.; Su, Y.l.; Li, P.; Wang, S. Isolation and characterization of Listeria monocytogenes in Chinese food obtained from the central area of China. Food Control 2017, 74, 9–16. [Google Scholar] [CrossRef]
- Li, C.; Zeng, H.; Ding, X.; Chen, Y.; Liu, X.; Zhou, L.; Wang, X.; Cheng, Y.; Hu, S.; Cao, Z.; et al. Perinatal listeriosis patients treated at a maternity hospital in Beijing, China, from 2013–2018. BMC Infect. Dis. 2020, 20, 601. [Google Scholar] [CrossRef]
- Andrews, S. Babraham Bioinformatics—FastQC A Quality Control tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 25 January 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 11 February 2022).
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. GitHub—Tseemann/mlst: Scan Contig Files against PubMLST Typing Schemes. Available online: https://github.com/tseemann/mlst (accessed on 11 February 2022).
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [Green Version]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]



| Virulence Gene | Allele | Virulence Gene | Allele |
|---|---|---|---|
| actA (lmo0204) | 120 | lisK (lmo1378) | 13 |
| agrA (lmo0051) | 6 | lisR (lmo1377) | 3 |
| agrC (lmo0050) | 12 (1 mismatch) * | lmo0333 (inlI) | 17 |
| ami (lmo2558) | 127 | lmo1280 (codY) | 3 |
| aut (lmo1076) | 6 | lmo2470 (inlP) | 9 |
| bsh (lmo2067) | 33 | lmo2491 (pdeE) | 2 |
| cheA (lmo0692) | 13 | lntA (lmo0438) | 48 |
| cheY (lmo0691) | 3 | lpeA (lmo1847) | 12 |
| cwhA (lmo0582; iap) | 15 | lplA1 (lmo0931) | 13 |
| dltA (lmo0974) | 13 | lspA (lmo1844) | 6 |
| fbpA (lmo1829) | 15 | mdrM (lmo1617) | 3 |
| fur (lmo1956) | 4 | mpl (lmo0203) | 16 |
| gtcA (lmo2549) | 7 | oatA (lmo1291) | 11 |
| hly (lmo0202) | 9 | oppA (lmo2196) | 8 |
| hpt (lmo0838) | 14 | pdgA (lmo0415) | 5 |
| inlA (lmo0433) | 12 | plcA (lmo0201) | 17 |
| inlB (lmo0434) | 15 | plcB (lmo0205) | 6 |
| inlC (lmo1786) | 13 | prfA (lmo0200) | 7 |
| inlC2 (LMON_RS01340) | 6 | prsA2 (lmo2219) | 5 |
| inlD (LMON_RS01345) | 6 | purQ (lmo1769) | 17 |
| inlE (lmo0264) | 4 | srtA (lmo0929) | 4 |
| inlF (lmo0409) | 10 | srtB (lmo2181) | 8 |
| inlH (lmo0263) | 12 | stp (lmo1821) | 5 |
| inlJ (lmo2821) | 172 | svpA (lmo2185) | 10 |
| inlK (lmo1290) | 14 | tagB (lmo1088) | 8 |
| lap (lmo1634) | 13 | vip (lmo0320) | 50 |
| lapB (lmo1666) | 11 | virR (lmo1745) | 4 |
| lgt (lmo2482) | 6 | virS (lmo1741) | 6 |
| ID Code | Sequenced Isolates | Food Sampling or Sampling Site | Sampling Date |
|---|---|---|---|
| L | Negative | Horse meat | 29 January 2020 |
| M | M1, M2 | Mortadella | 5 February 2020 |
| E | E1, E2, E3 | First meat slicer | 6 February 2020 |
| F | Negative | Worktable | 6 February 2020 |
| G | G1, G2 | Second meat slicer | 6 February 2020 |
| H | H1, H2 | Cold food counter | 6 February 2020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russini, V.; Spaziante, M.; Varcasia, B.M.; Diaconu, E.L.; Paolillo, P.; Picone, S.; Brunetti, G.; Mattia, D.; De Carolis, A.; Vairo, F.; et al. A Whole Genome Sequencing-Based Epidemiological Investigation of a Pregnancy-Related Invasive Listeriosis Case in Central Italy. Pathogens 2022, 11, 667. https://doi.org/10.3390/pathogens11060667
Russini V, Spaziante M, Varcasia BM, Diaconu EL, Paolillo P, Picone S, Brunetti G, Mattia D, De Carolis A, Vairo F, et al. A Whole Genome Sequencing-Based Epidemiological Investigation of a Pregnancy-Related Invasive Listeriosis Case in Central Italy. Pathogens. 2022; 11(6):667. https://doi.org/10.3390/pathogens11060667
Chicago/Turabian StyleRussini, Valeria, Martina Spaziante, Bianca Maria Varcasia, Elena Lavinia Diaconu, Piermichele Paolillo, Simonetta Picone, Grazia Brunetti, Daniela Mattia, Angela De Carolis, Francesco Vairo, and et al. 2022. "A Whole Genome Sequencing-Based Epidemiological Investigation of a Pregnancy-Related Invasive Listeriosis Case in Central Italy" Pathogens 11, no. 6: 667. https://doi.org/10.3390/pathogens11060667






