Exogenous Rubella Virus Capsid Proteins Enhance Virus Genome Replication
Abstract
:1. Introduction
2. Results
2.1. Exogenous Virion Proteins Enhance RuV Genome Replication
2.2. The RuV CP Is the Major Component Enhancing RuV Replication
2.3. RuV CP Does Not Affect Protein Synthesis
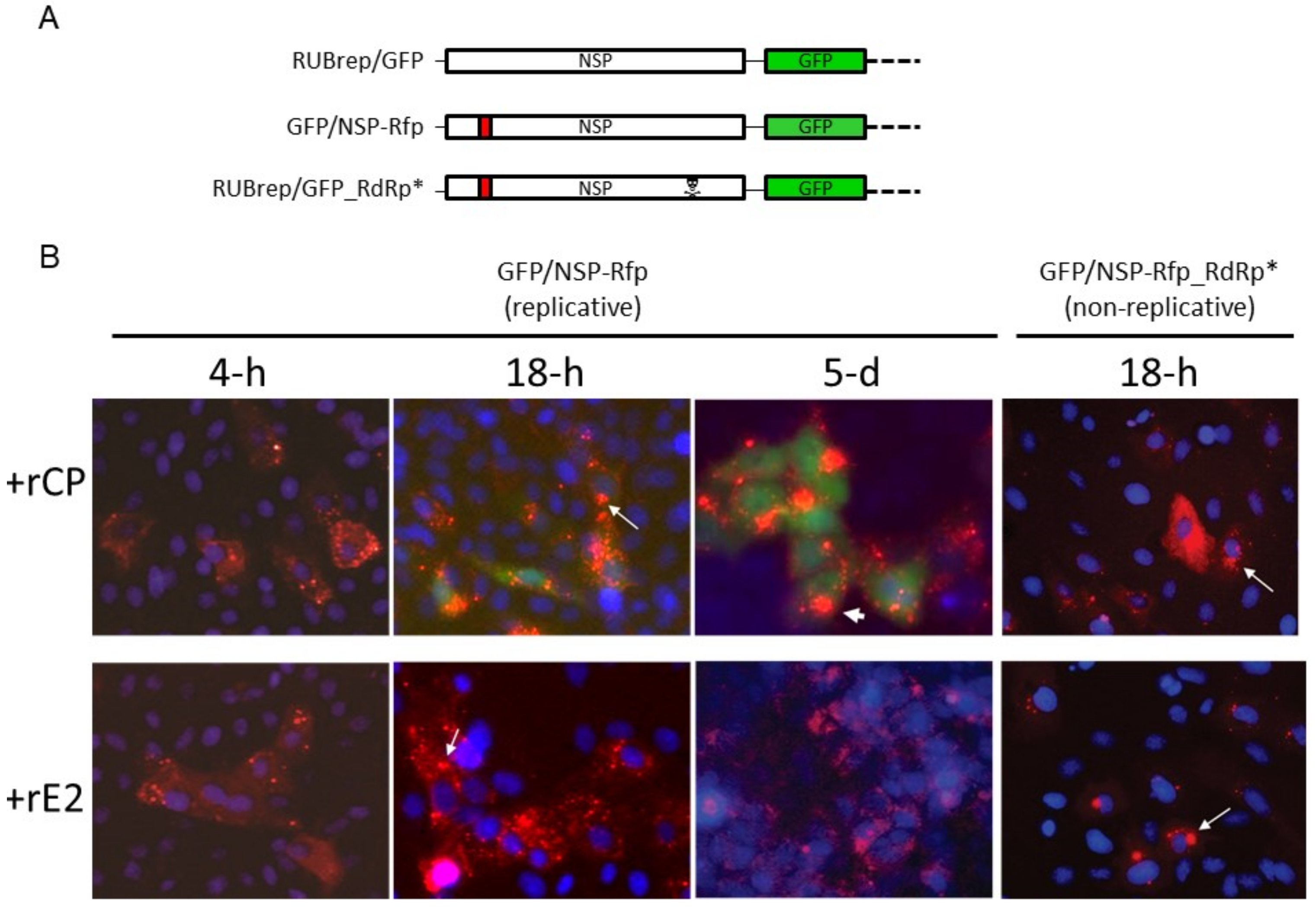
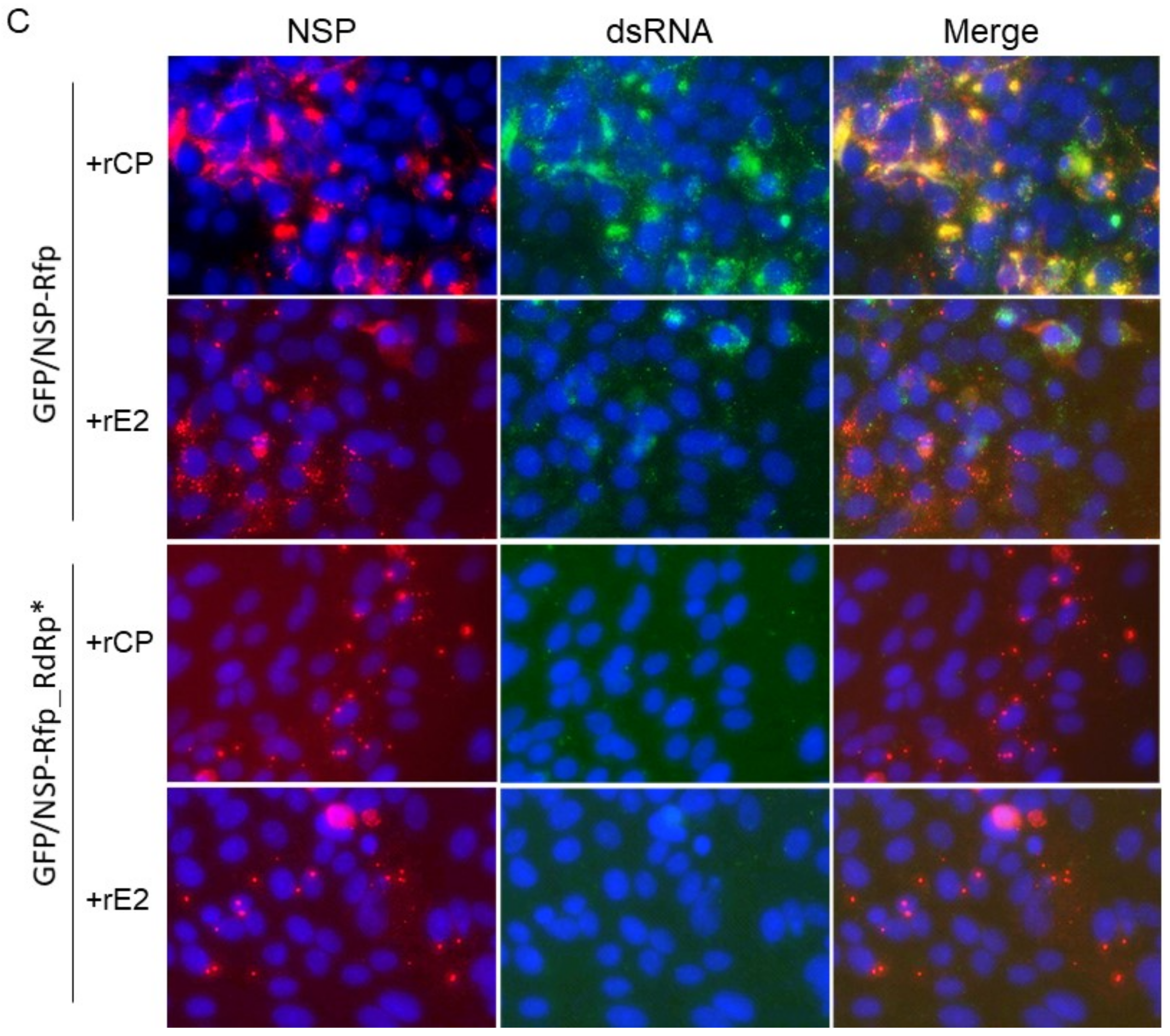
2.4. Exogenous RuV CP Does Not Affect Intracellular Localization of RuV NSP
2.5. RuV CP Affects the gRNA, but Not sgRNA Synthesis
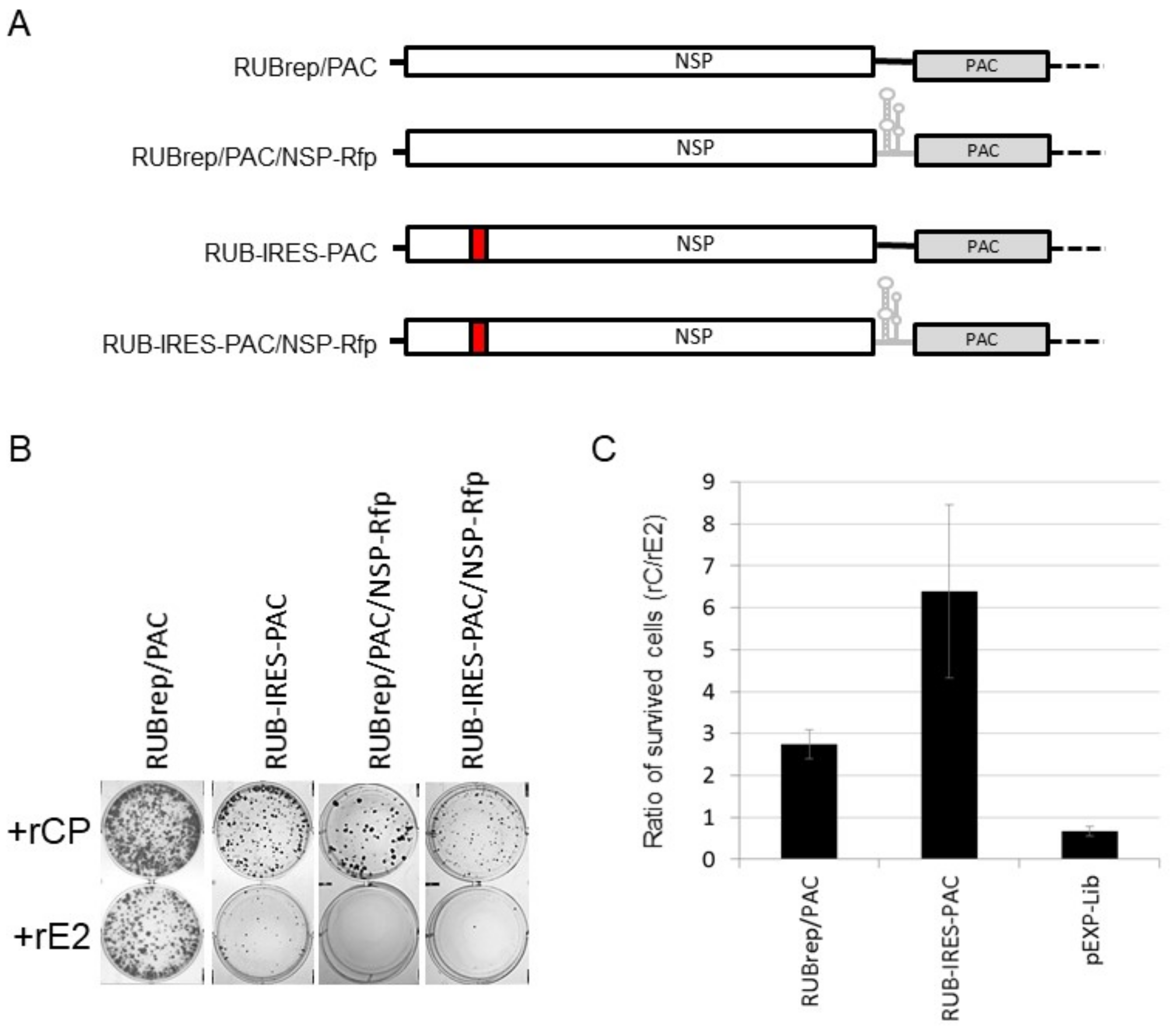
2.6. RuV CP Rescues the Replication of a Spectrum of RuV Mutants and Affects RNA Stability
3. Discussion
4. Materials and Methods
4.1. Cell Culture, Virus Preparation, and Titration
4.2. Virus Antigens and Pseudovirus
4.3. Constructs and Transfection
4.4. In Vitro Transcription and Translation
4.5. Northern Hybridization and RT-PCR
4.6. Quantification of Reporter Proteins and RNA
4.7. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Chen, R.; Mukhopadhyay, S.; Merits, A.; Bolling, B.; Nasar, F.; Coffey, L.L.; Powers, A.; Weaver, S.C.; Ictv Report Consortium. ICTV Virus Taxonomy Profile: Togaviridae. J. Gen. Virol. 2018, 99, 761–762. [Google Scholar] [CrossRef] [PubMed]
- Abernathy, E.; Chen, M.-H.; Bera, J.; Shrivastava, S.; Kirkness, E.; Zheng, Q.; Bellini, W.; Icenogle, J.P. Analysis of whole genome sequences of 16 strains of rubella virus from the United States, 1961–2009. Virol. J. 2013, 10, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.-H.; Icenogle, J.P. Molecular virology of rubella virus. In Rubella Viruses, 1st ed.; Banatvala, J., Peckham, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; Volume 15, pp. 1–18. [Google Scholar]
- Liang, Y.; Yao, J.; Gillam, S. Rubella virus nonstructural prostein protease domains involved in trans- and cis-cleavage activities. J. Virol. 2000, 74, 5412–5423. [Google Scholar] [CrossRef] [Green Version]
- Gros, G.; Wengler, G. Identification of an RNA-stimulated NTPase in the predicted helicase sequence of the Rubella virus nonstructural polyprotein. Virology 1996, 217, 367–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Gillam, S. Mutations in the GDD motif of rubella virus putative RNA-dependent RNA polymerase affect virus replication. Virology 2001, 285, 322–331. [Google Scholar] [CrossRef] [Green Version]
- Qiu, Z.; Ou, D.; Hobman, T.C.; Gillam, S. Expression and characterization of virus-like particles containing rubella virus structural proteins. J. Virol. 1994, 68, 4086–4091. [Google Scholar] [CrossRef] [Green Version]
- Oker-Blom, C.; Ulmanen, I.; Kääriäinen, L.; Pettersson, R.F. Rubella virus 40 S genome RNA specifies a 24 S subgenomic mRNA that codes for a precursor to structural proteins. J. Virol. 1984, 49, 403–408. [Google Scholar] [CrossRef] [Green Version]
- Pugachev, K.V.; Abernathy, E.S.; Frey, T.K. Improvement of the specific infectivity of the rubella virus (RUB) infectious clone: Determinants of cytopathogenicity induced by RUB map to the nonstructural proteins. J. Virol. 1997, 71, 562–568. [Google Scholar] [CrossRef] [Green Version]
- Tzeng, W.P.; Chen, M.-H.; Derdeyn, C.A.; Frey, T.K. Rubella virus DI RNAs and replicons: Requirement for nonstructural proteins acting in cis for amplification by helper virus. Virology 2001, 289, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Frolov, I.; Icenogle, J.P.; Frey, T.K. Analysis of the 3′ cis-acting elements of rubella virus by using replicons expressing a puromycin resistance gene. J. Virol. 2004, 78, 2553–2561. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Icenogle, J.P. Rubella virus capsid protein modulates viral genome replication and virus infectivity. J. Virol. 2004, 78, 4314–4322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzeng, W.P.; Frey, T.K. Complementation of a deletion in the rubella virus p150 nonstructural protein by the viral capsid protein. J. Virol. 2003, 77, 9502–9510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzeng, W.P.; Frey, T.K. Functional replacement of a domain in the rubella virus p150 replicase protein by the virus capsid protein. J. Virol. 2009, 83, 3549–3555. [Google Scholar] [CrossRef] [Green Version]
- Tzeng, W.P.; Matthews, J.D.; Frey, T.K. Analysis of rubella virus capsid protein-mediated enhancement of replicon replication and mutant rescue. J. Virol. 2006, 80, 3966–3974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, V.M.; Willows, S.D.; Fokine, A.; Battisti, A.J.; Sun, S.; Plevka, P.; Hobman, T.C.; Rossmann, M.G. Rubella virus capsid protein structure and its role in virus assembly and infection. Proc. Natl. Acad. Sci. USA 2013, 110, 20105–20110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, L.M.; Everitt, J.C.; Beatch, M.D.; Holmes, C.F.; Hobman, T.C. Phosphorylation of rubella virus capsid regulates its RNA binding activity and virus replication. J. Virol. 2003, 77, 1764–1771. [Google Scholar] [CrossRef] [Green Version]
- Law, L.J.; Ilkow, C.S.; Tzeng, W.P.; Rawluk, M.; Stuart, D.T.; Frey, T.K.; Hobman, T.C. Analyses of phosphorylation events in the rubella virus capsid protein: Role in early replication events. J. Virol. 2006, 80, 6917–6925. [Google Scholar] [CrossRef] [Green Version]
- Singh, I.; Helenius, A. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol. 1992, 66, 7049–7058. [Google Scholar] [CrossRef] [Green Version]
- Wengler, G.; Wengler, G. Identification of a transfer of viral core protein to cellular ribosomes during the early stages of alphavirus infection. Virology 1984, 134, 435–442. [Google Scholar] [CrossRef]
- Battisti, A.J.; Yoder, J.D.; Plevka, P.; Winkler, D.C.; Prasad, V.M.; Kuhn, R.J.; Frey, T.K.; Steven, A.C.; Rossmann, M.G. Cryo-electron tomography of rubella virus. J. Virol. 2012, 86, 11078–11085. [Google Scholar] [CrossRef] [Green Version]
- Bennett, A.J.; Paskey, A.C.; Ebinger, A.; Pfaff, F.; Priemer, G.; Höper, D.; Breithaupt, A.; Heuser, E.; Ulrich, R.G.; Kuhn, J.H.; et al. Relatives of rubella virus in diverse mammals. Nature 2020, 586, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Kujala, P.; Ahola, T.; Ehsani, N.; Auvinen, P.; Vihinen, H.; Kääriäinen, L. Intracellular distribution of rubella virus nonstructural protein P150. J. Virol. 1999, 73, 7805–7811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, J.; Tzeng, W.P.; Calderita, G.; Fraile-Ramos, A.; Frey, T.K.; Risco, C. Novel replication complex architecture in rubella replicon-transfected cells. Cell Microbiol. 2007, 9, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Marshall, J.A.; Bowden, D.S. Localization of rubella virus core particles in Vero cells. Virology 1999, 265, 110–119. [Google Scholar] [CrossRef] [Green Version]
- Tzeng, W.P.; Zhou, Y.; Icenogle, J.P.; Frey, T.K. Novel replicon-based reporter gene assay for detection of rubella virus in clinical specimens. J. Clin. Microbiol. 2005, 43, 879–885. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Xu, W.; Abernathy, E.S.; Chen, M.H.; Zheng, Q.; Wang, T.; Zhang, Z.; Li, C.; Wang, C.; He, W.; et al. Comparison of four methods using throat swabs to confirm rubella virus infection. J. Clin. Microbiol. 2007, 45, 2847–2852. [Google Scholar] [CrossRef] [Green Version]
- Burns, C.C.; Shaw, J.; Campagnoli, R.; Jorba, J.; Vincent, A.; Quay, J.; Kew, O. Modulation of Poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J. Virol. 2006, 80, 3259–3272. [Google Scholar] [CrossRef] [Green Version]
- Matthews, J.D.; Tzeng, W.P.; Frey, T.K. Determinants in the maturation of the rubella virus P200 replicase polyprotein precursor. J. Virol. 2012, 86, 6457–6469. [Google Scholar] [CrossRef] [Green Version]
- Forng, R.Y.; Frey, T.K. Identification of the rubella virus nonstructural proteins. Virology 1995, 206, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Gillam, S. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J. Virol. 2000, 74, 5133–5141. [Google Scholar] [CrossRef]
- Tzeng, W.P.; Frey, T.K. Mapping the rubella virus subgenomic promoter. J. Virol. 2002, 76, 3189–3201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pugachev, K.V.; Tzeng, W.P.; Frey, T.K. Development of a rubella virus vaccine expression vector: Use of a picornavirus internal ribosome entry site increases stability of expression. J. Virol. 2000, 74, 10811–10815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hemphill, M.L.; Forng, R.Y.; Abernathy, E.S.; Frey, T.K. Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology 1988, 162, 65–75. [Google Scholar] [CrossRef]
- Tzeng, W.P.; Frey, T.K. Rubella virus capsid protein modulation of viral genomic and subgenomic RNA synthesis. Virology 2005, 337, 327–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilkow, C.S.; Willows, S.D.; Hobman, T.C. Rubella virus capsid protein: A small protein with big functions. Future Microbiol. 2010, 5, 571–584. [Google Scholar] [CrossRef]
- Ilkow, C.S.; Mancinelli, V.; Beatch, M.D.; Hobman, T.C. Rubella virus capsid protein interacts with poly(a)-binding protein and inhibits translation. J. Virol. 2008, 82, 4284–4294. [Google Scholar] [CrossRef] [Green Version]
- Ilkow, C.S.; Weckbecker, D.; Cho, W.J.; Meier, S.; Beatch, M.D.; Goping, I.S.; Herrmann, J.M.; Hobman, T.C. The rubella virus capsid protein inhibits mitochondrial import. J. Virol. 2010, 84, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Ilkow, C.S.; Goping, I.S.; Hobman, T.C. The Rubella virus capsid is an anti-apoptotic protein that attenuates the pore-forming ability of Bax. PLoS Pathog. 2011, 7, e1001291. [Google Scholar] [CrossRef] [Green Version]
- Duncan, R.; Esmaili, A.; Law, L.M.; Bertholet, S.; Hough, C.; Hobman, T.C.; Nakhasi, H.L. Rubella virus capsid protein induces apoptosis in transfected RK13 cells. Virology 2000, 275, 20–29. [Google Scholar] [CrossRef] [Green Version]
- Willows, S.; Ilkow, C.S.; Hobman, T.C. Phosphorylation and membrane association of the Rubella virus capsid protein is important for its anti-apoptotic function. Cell Microbiol. 2014, 16, 1201–1210. [Google Scholar] [CrossRef]
- Claus, C.; Tzeng, W.P.; Liebert, U.; Frey, T.K. Rubella virus-like replicon particles: Analysis of encapsidation determinants and non-structural roles of capsid protein in early post-entry replication. J. Gen. Virol. 2012, 93, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Makino, S. Interplay between viruses and host mRNA degradation. Biochim. Biophys. Acta 2013, 1829, 732–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, S.D.; Tzeng, W.P.; Chen, M.-H.; Frey, T.K. Analysis of intermolecular RNA-RNA recombination by rubella virus. Virology 2003, 309, 258–271. [Google Scholar] [CrossRef] [Green Version]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.-H.; Frey, T.K. Mutagenic analysis of the 3′ cis-acting elements of the rubella virus genome. J. Virol. 1999, 73, 3386–3403. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.-H.; Zhu, Z.; Zhang, Y.; Favors, S.; Xu, W.B.; Featherstone, D.A.; Icenogle, J.P. An indirect immunocolorimetric assay to detect rubella virus infected cells. J. Virol. Methods 2007, 146, 414–418. [Google Scholar] [CrossRef]
- Jan, J.T.; Griffin, D.E. Induction of Apoptosis by Sindbis Virus Occurs at Cell Entry and Does Not Require Virus Replication. J. Virol. 1999, 73, 10296–10302. [Google Scholar] [CrossRef] [Green Version]
- Klein, D.; Bugl, B.; Günzburg, W.H.; Salmons, B. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gene Ther. 2000, 7, 458–463. [Google Scholar] [CrossRef] [Green Version]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]



| SINV | PV | RuV | LacZ Control | |
|---|---|---|---|---|
| Fold change in titer or activity (+VLP/−VLP) | 0.14 ± 0.08 | 0.08 ± 0.06 | 2.99 ± 1.62 | 0.91 ± 0.28 |
| Mutant | Description | Domain | GFP Expression a | References | |
|---|---|---|---|---|---|
| +rCP | +rE2 | ||||
| ATG* | Mutate NSP ORF start codon to TAG | − | − | − | This study |
| ΔNotI | Delete nt 1694–2191 (aa 552–717) from NSP | In P150; within Q domain | ++ | − | [10,14] |
| NSP-Rfp | Replace nt 1694–2191 with RFP gene | In P150; within Q domain | ++ | − | This study |
| 1152S | Mutate Cys1152 to Ser | In P150; catalytic domain of viral protease | + 2 | − | [31] |
| 1301S | Mutate Gly1301 to Ser | Cleavage site by RuV protease | + | − | [31] |
| GA205D | Mutate Asp 205 to Ala | In p150; unknown domain | − | − | This study |
| GA1326D | Mutate Asp 1326 to Ala | In P90; unknown | + 2 | − | This study |
| GA1967D | Mutate Asp 1967 to Ala | In p90; putative RdRp catalytic domain | ++ | − | [6] |
| RdRp* (GK1967L1968) | Mutate Asp 1967 to Lys and Asp 1968 to Leu | In P90; putative RdRp catalytic domain | − | − | This study |
| P90-His | Add six histidine (His) residues at the C-terminus of p90 | In p90; unknown | ++ | − | This study |
| Constructs | Description | Mutagenesis | Refs. | |
|---|---|---|---|---|
| RuV infectious clone | Robo402 | RuV infectious cDNA clone | ND (not needed) | [9] |
| Robo402ires | RuV infectious cDNA clone with the intergenic region replaced with an internal ribosomal entry site (IRES) of encephalomyocarditis virus (EMCV) | ND | ||
| RuV replicons | RUBrep/GFP | RuV replicon with partial SP coding region replaced by green fluorescent protein (GFP) gene | ND | [10] |
| RUBrep/Rfp | RuV replicon with partial SP coding region (nt 6512 to 9333) replaced by red fluorescent protein (RFP) gene | PCR amplified RFP gene and swapped with the Xba I-Nsi I fragment in RUBrep/GFP | ||
| RUBrep/PAC | RuV replicon with partial SP coding region (nt 6512 to 9179) replaced by puromycin-N-acetyltransferase (PAC) | ND | [12] | |
| RUBrep/GFP_ΔNotI | RUBrep/GFP with nt 1693 to 2191 deleted | ND | [10] | |
| RUBrep/GFP/NSP-Rfp | RUBrep/GFP with RFP gene inserted between nt 1693 and 2191 | PCR amplified RFP gene and swapped with the Not I region in RUBrep/GFP | ||
| RUBrep/PAC/NSP-Rfp | RUBrep/PAC with RFP gene inserted between nt 1693 and 2191 | PCR amplified RFP gene and swapped with the Not I region in RUBrep/PAC | ||
| RUB-IRES-PAC | RUBrep/PAC replicon with the intergenic region (nt 6392–6511; IR) replaced by the EMCV IRES | PCR amplified partial NSP and IRES element of Robo402ires and swapped with the Fse I and Xba I fragment in RUBrep/PAC | ||
| RUB-IRES-PAC/NSP-Rfp | RUBrep/PAC/NSP-Rfp replicon with IR replaced by EMCV IRES element | PCR amplified partial NSP and IRES element of Robo402ires and swapped with the Fse I and Xba I fragment in RUBrep/PAC/NSP-Rfp | ||
| RUBrep/GFP_NSP-ATG* | RUBrep/GFP replicons with changes in the start codon ATG to TAG | PCR amplification with mutagenic primers and swapped the HindIII-Bsu36I fragment (nt 1–499) | ||
| RUBrep/GFP_1152S | RUBrep/GFP replicons with a single mutation in the catalytic pocket of RuV nonstructural protease at Cys1152 to Ser | PCR amplification with mutagenic primers and swapped the Bsu36 II-Cla I fragment (nt 499–4392) | [31] | |
| RUBrep/GFP_1301S | RUBrep/GFP replicons with the cleavage site (Gly 1301) of nonstructural polyprotein mutated to Ser | PCR amplification with mutagenic primers and swapped the Bsu36 II-Cla I fragment (nt 499–4392) | [31] | |
| RUBrep/GFP_RdRp* | RUBrep/GFP replicons with changes in the putative RNA-dependent RNA polymerase catalytic domain at Asp1967 to Lys and Asp1968 to Leu | PCR amplification with mutagenic primers and swapped the Bgl II-Fse I fragment (nt 5355–6091) | ||
| RuV mini-Xpress system | g41-GFP (or g41-Rfp) | RuV genomic mini replicon with RuV 5′ 41-nt fused with the GFP (or RFP) gene followed by 3′ terminal 400 nts (or 600 nts for g41-Rfp) | Replacing the EcoN I-EcoRI fragment from Robo402 with the PCR amplified subgenomic region of RUBrep/GFP (or RUBrep/Rfp) (with EcoN I-EcoR I sites) | |
| g1700-GFP (or g1700-Rfp) | RuV expression system RuV 5′1692-nt fused with the GFP (or RFP) gene followed by 3′ terminal 400 nts (or 600 nts for g1700-Rfp) | Replacing the Not I-EcoRI fragment from Robo402 with the PCR amplified subgenomic region of RUBrep/GFP (or RUBrep/Rfp) (with Not I-EcoR I sites) | ||
| pUC-sg-GFP (or pUC-sg-Rfp) | RuV expression system containing the subgenomic sequences of RUBrep/GFP (or RUBrep/Rfp) | PCR amplification of subgenomic RNA sequences from RUBrep/GFP (or RUBrep/Rfp) and clone to pUC18 vector; the forward primer contains Hind III site and SP6 RNA polymerase promoter | ||
| Controls | pCI-GFP (or RFP) | GFP (or RFP) control plasmid in pCI-Neo vector (Promega) | PCR amplified GFP (or RFP) gene was cloned to pCI-Neo vector (Promega) between Nhe I-EcoR I sites | |
| pGEM-GFP | PCR controls or probe syntheses; in pGEM3Zf(−) vector (Promega) | PCR amplified GFP gene was cloned to pGEM3Zf(−) vector (Promega) between HindIII-EcoR I sites |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.-H.; Burns, C.C.; Abernathy, E.; Ogee-Nwankwo, A.A.; Icenogle, J.P. Exogenous Rubella Virus Capsid Proteins Enhance Virus Genome Replication. Pathogens 2022, 11, 683. https://doi.org/10.3390/pathogens11060683
Chen M-H, Burns CC, Abernathy E, Ogee-Nwankwo AA, Icenogle JP. Exogenous Rubella Virus Capsid Proteins Enhance Virus Genome Replication. Pathogens. 2022; 11(6):683. https://doi.org/10.3390/pathogens11060683
Chicago/Turabian StyleChen, Min-Hsin, Cara C. Burns, Emily Abernathy, Adaeze A. Ogee-Nwankwo, and Joseph P. Icenogle. 2022. "Exogenous Rubella Virus Capsid Proteins Enhance Virus Genome Replication" Pathogens 11, no. 6: 683. https://doi.org/10.3390/pathogens11060683
APA StyleChen, M.-H., Burns, C. C., Abernathy, E., Ogee-Nwankwo, A. A., & Icenogle, J. P. (2022). Exogenous Rubella Virus Capsid Proteins Enhance Virus Genome Replication. Pathogens, 11(6), 683. https://doi.org/10.3390/pathogens11060683






