T-Cell Receptor β Chain and B-Cell Receptor Repertoires in Chronic Hepatitis B Patients with Coexisting HBsAg and Anti-HBs
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
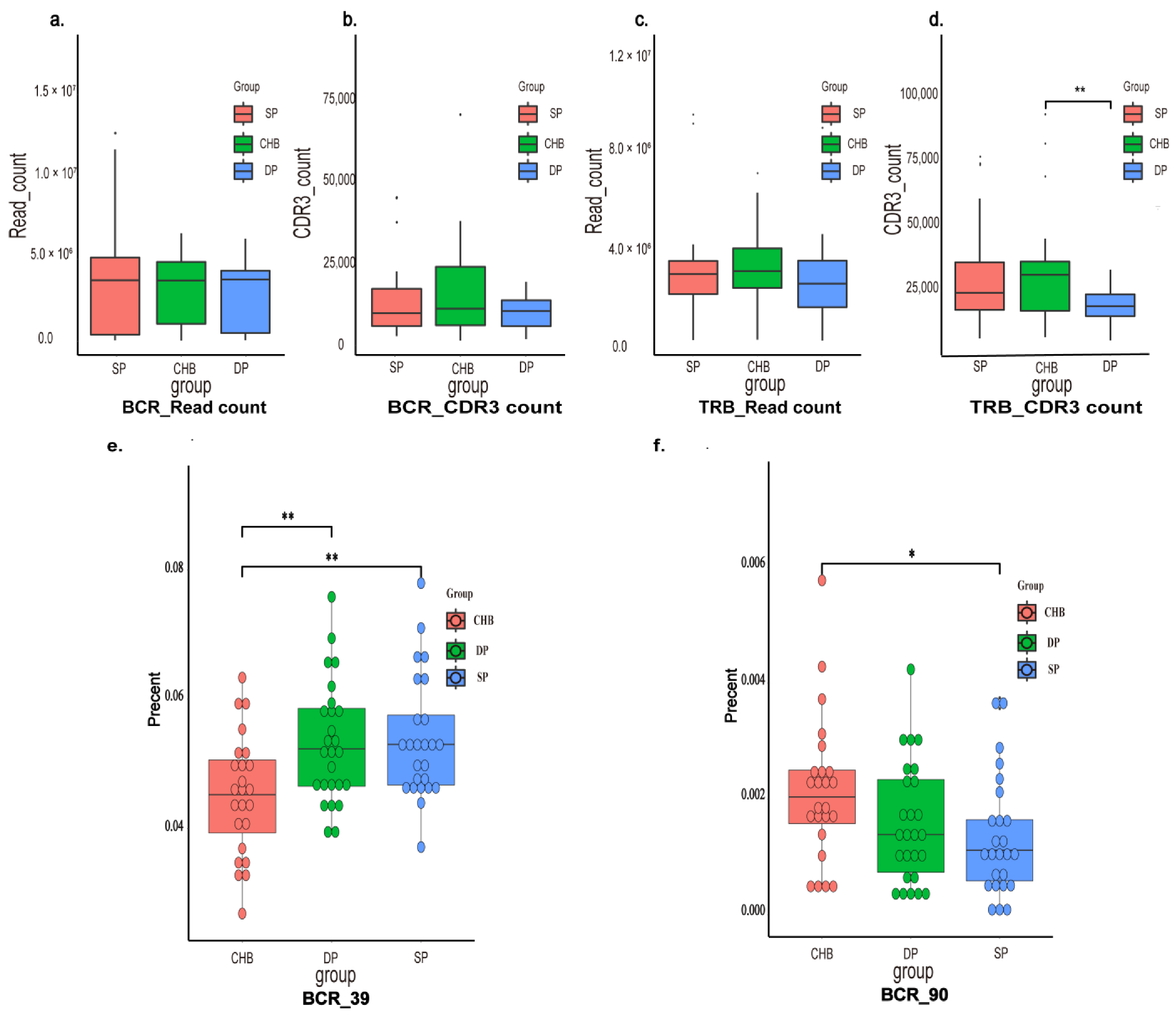
2.2. Immune Repertoire Profiling
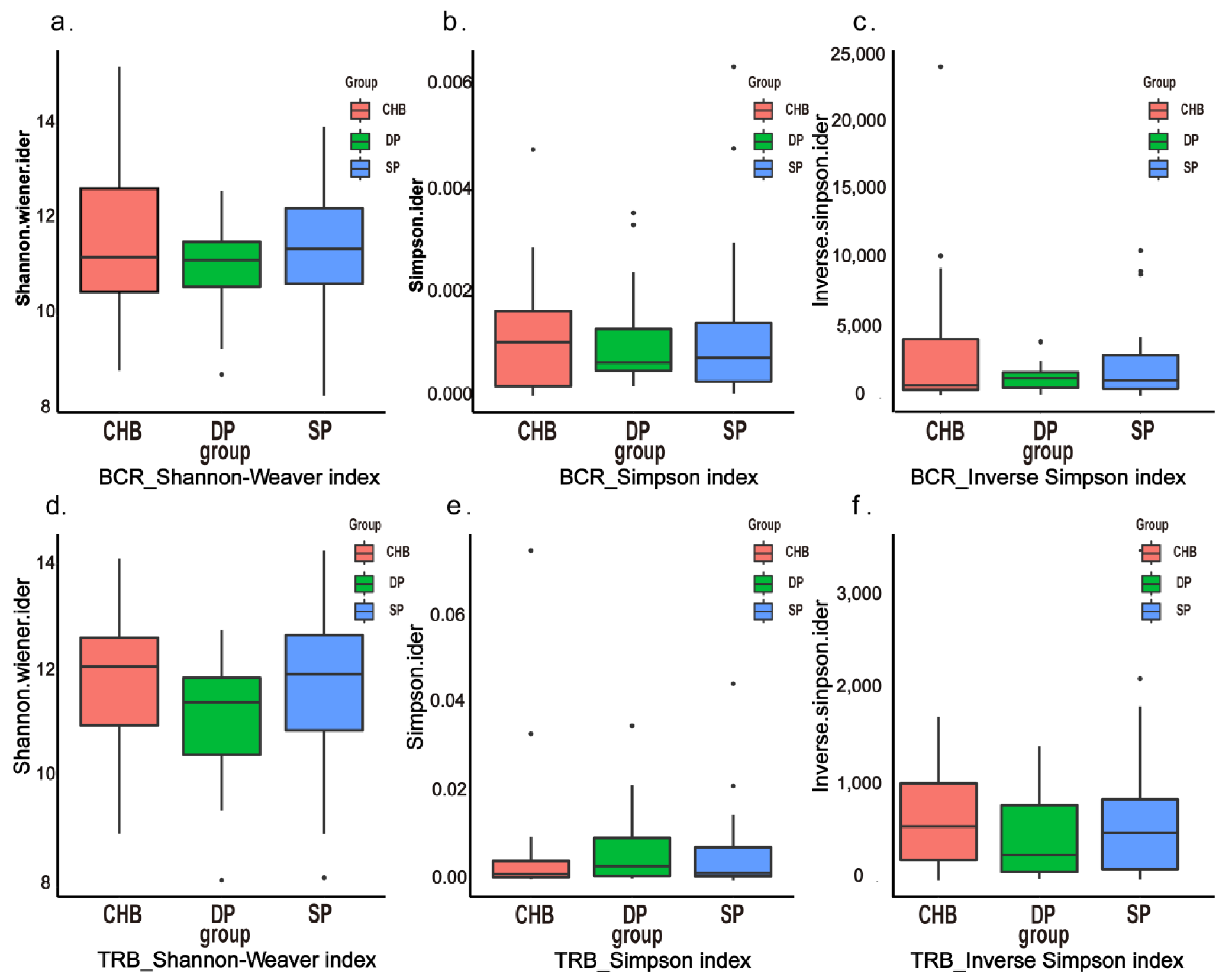
2.3. Diversity Analysis
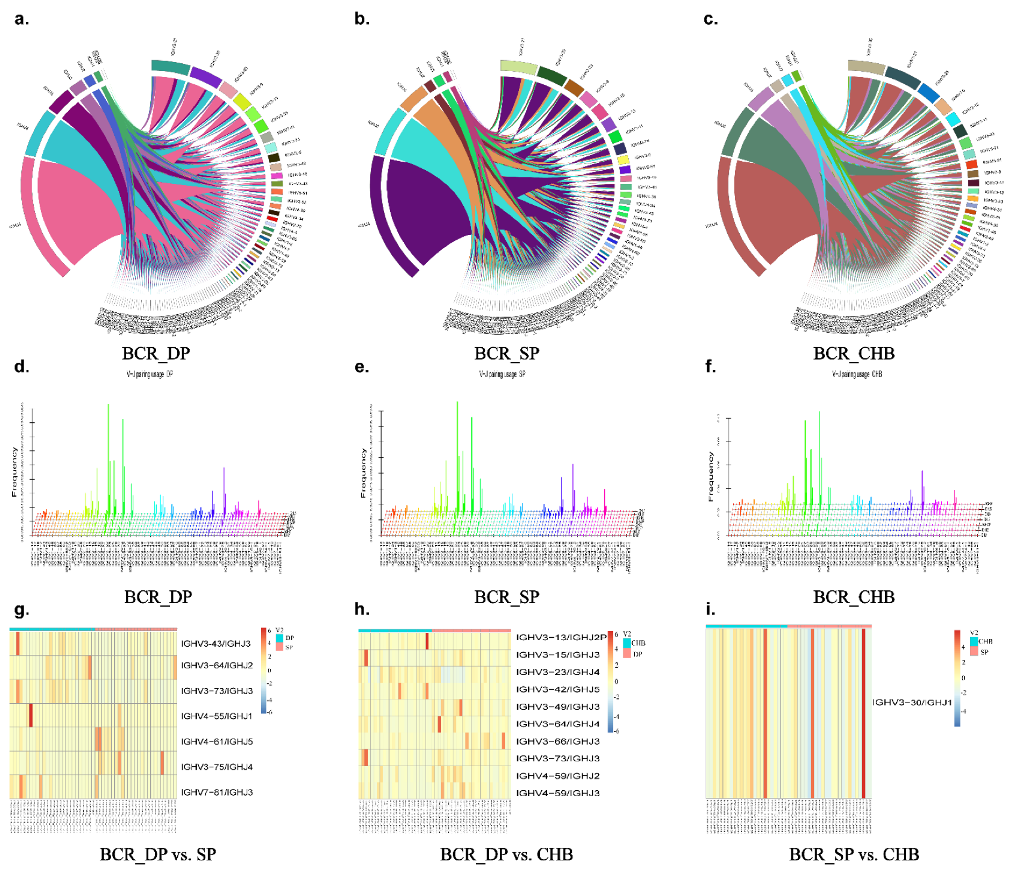
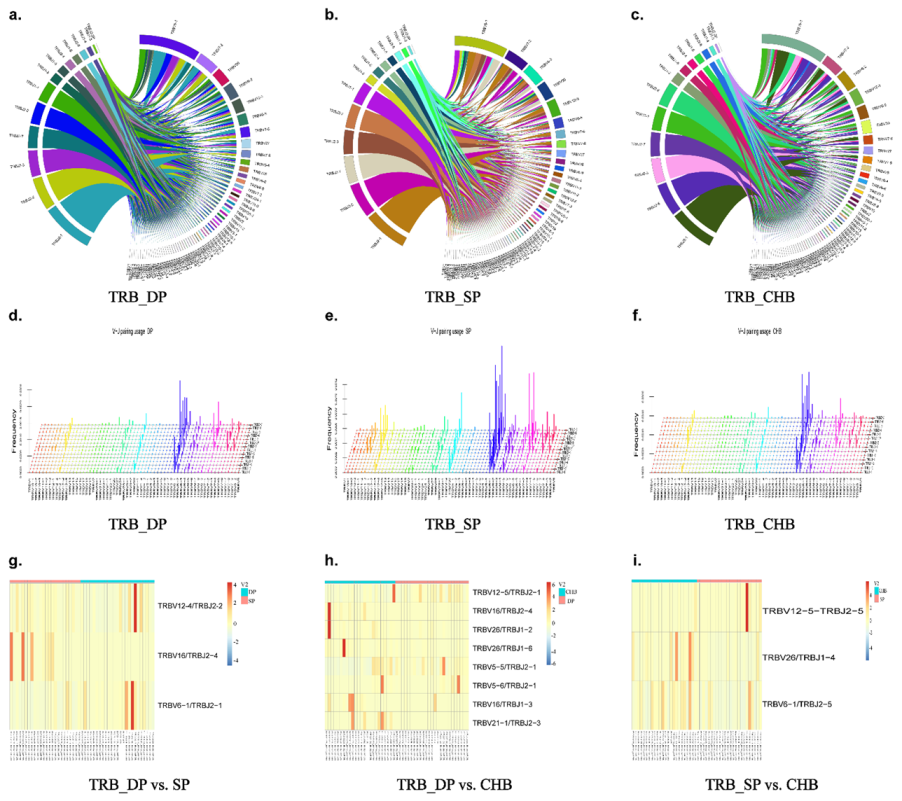
2.4. Analysis of V, D, and J Gene Segments
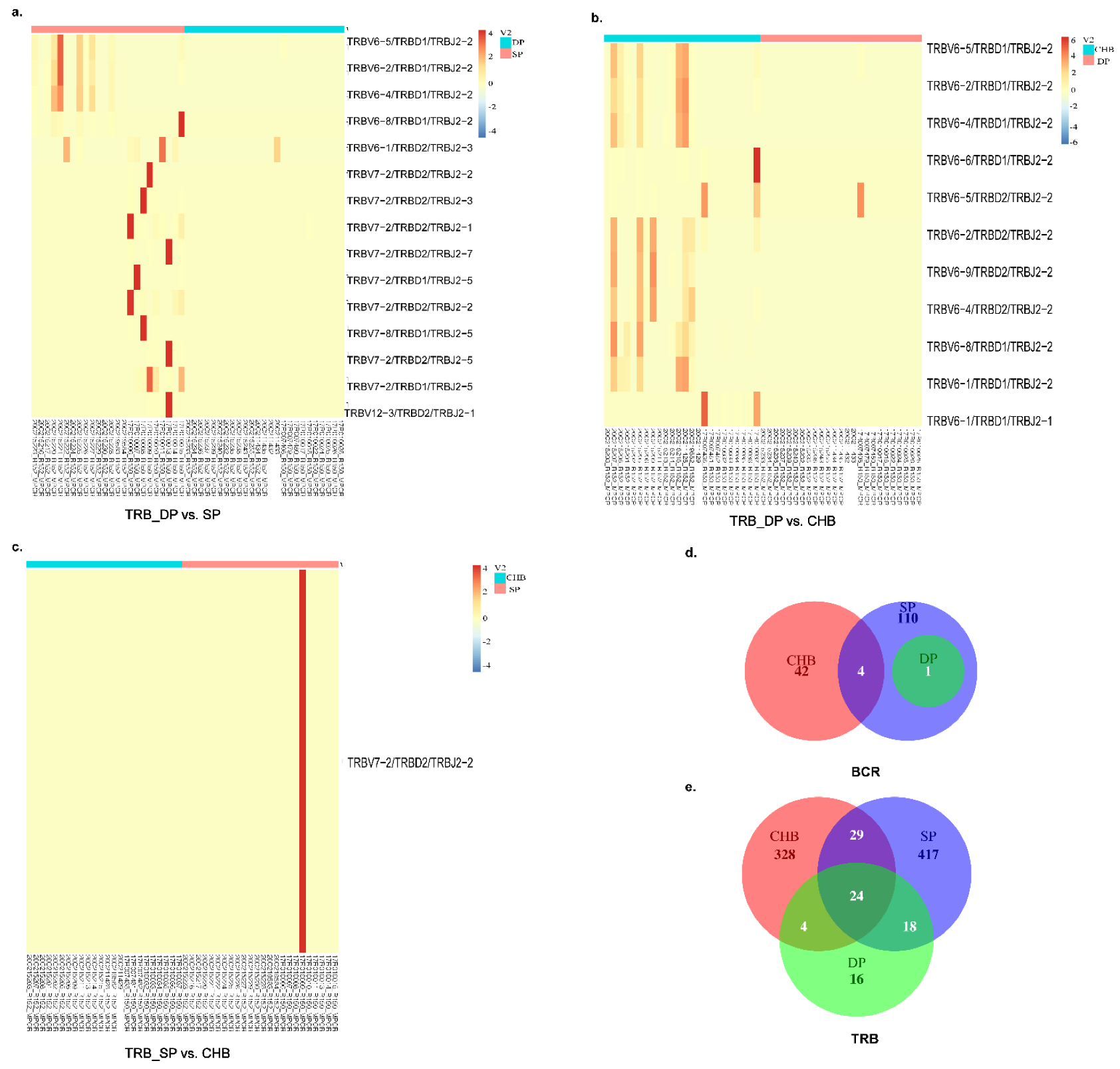
2.5. Analysis of V–J, V–D–J Gene Combinations, and Amino Acid Clonotypes
2.6. Principal Component Analysis
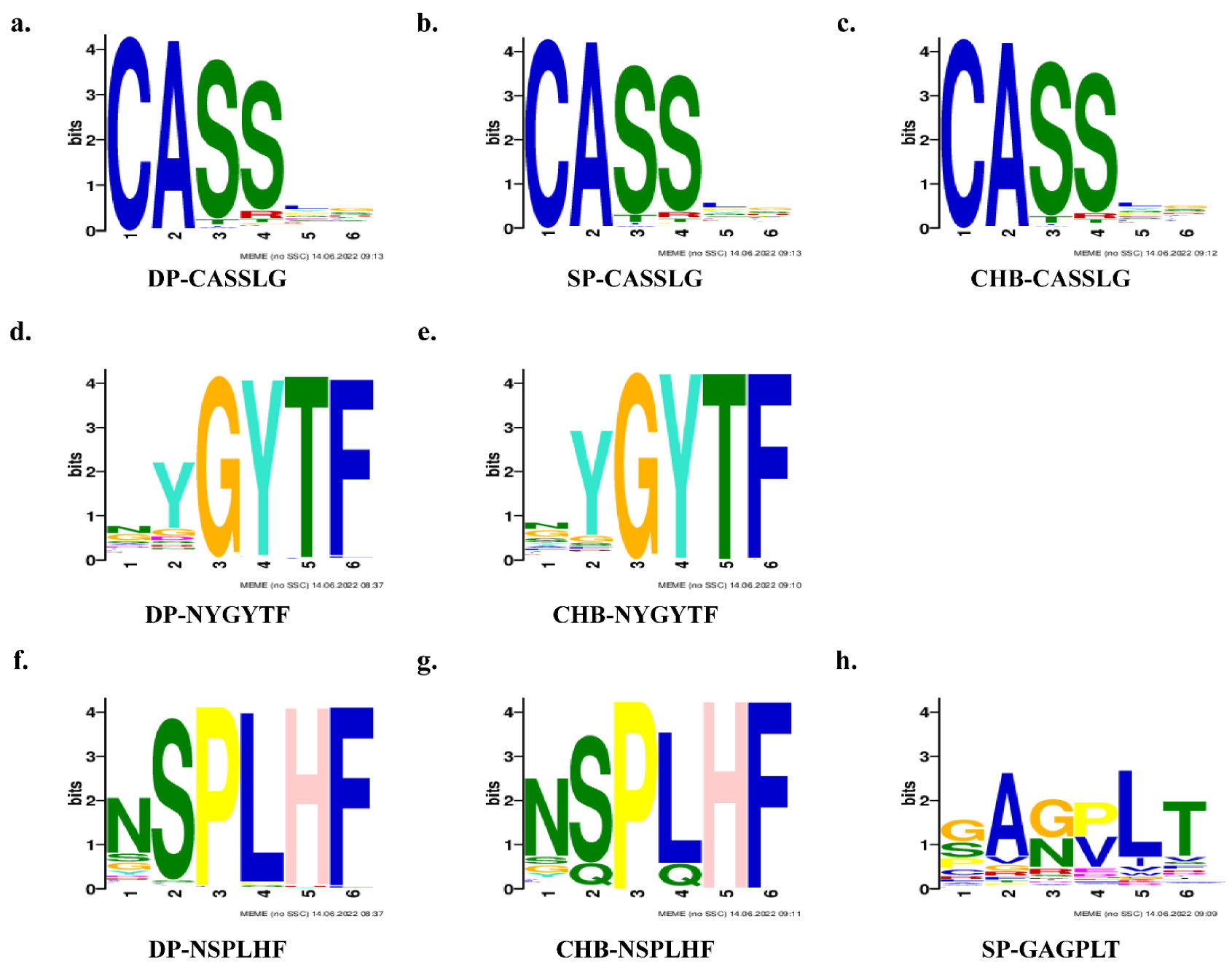
2.7. TCR Clusters Analysis and Motifs Prediction
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. DNA Extraction and Immune Repertoire Library Construction
4.3. Data Processing and Sequence Analysis
4.4. Clustering Estimation and Motifs Prediction of TRB Clonotypes
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Hepatitis B. Available online: www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed on 1 September 2021).
- Paik, J.M.; Younossi, Y.; Henry, L.; Mishra, A.; Younossi, Z.M. Recent trends in the global burden of hepatitis B virus: 2007–2017. Gastroenterology 2021, 160, P1845–P1846.E3. [Google Scholar] [CrossRef] [PubMed]
- Arnold, W.; Hess, G.; Meyer zum Büschenfelde, K.H.; Kösters, W.; Biswas, R.; Strauch, M.; Knolle, J. Simultaneous occurrence in the serum of hepatitis B surface antigen (HBsAg) and antibodies against HBsAg (Anti-HBs) of different subtypes. (Serologic and fluorescence histologic studies). Verh. Dtsch. Ges. Inn. Med. 1976, 82, 405–407. [Google Scholar] [PubMed]
- Jiang, X.; Chang, L.; Yan, Y.; Wang, L. Paradoxical HBsAg and anti-HBs coexistence among chronic HBV infections: Causes and consequences. Int. J. Biol. Sci. 2021, 17, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.N.; Nokes, D.J.; Carman, W.F. Current status of HBV vaccine escape variants--a mathematical model of their epidemiology. J. Viral. Hepat. 1998, 5 (Suppl. S2), 25–30. [Google Scholar] [CrossRef]
- Alavian, S.M.; Carman, W.F.; Jazayeri, S.M. HBsAg variants: Diagnostic-escape and diagnostic dilemma. J. Clin. Virol. 2013, 57, 201–208. [Google Scholar] [CrossRef]
- Günther, S.; Fischer, L.; Pult, I.; Sterneck, M.; Will, H. Naturally occurring variants of hepatitis B virus. Adv. Virus Res. 1999, 52, 25–137. [Google Scholar] [CrossRef]
- Lada, O.; Benhamou, Y.; Poynard, T.; Thibault, V. Coexistence of hepatitis B surface antigen (HBsAg) and anti-HBs antibodies in chronic hepatitis B virus carriers: Influence of “a” determinant variants. J. Virol. 2006, 80, 2968–2975. [Google Scholar] [CrossRef] [Green Version]
- Colson, P.; Borentain, P.; Motte, A.; Henry, M.; Moal, V.; Botta-Fridlund, D.; Tamalet, C.; Gérolami, R. Clinical and virological significance of the co-existence of HBsAg and anti-HBs antibodies in hepatitis B chronic carriers. Virology 2007, 367, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Ding, F.; Miao, X.-L.; Li, Y.-X.; Dai, J.-F.; Yu, H.-G. Mutations in the S gene and in the overlapping reverse transcriptase region in chronic hepatitis B Chinese patients with coexistence of HBsAg and anti-HBs. Braz. J. Infect. Dis. 2016, 20, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.-C.; Li, X.; Li, L.; Li, X.-F.; Zhang, L.; Wei, J. Evolution of full-length genomes of HBV quasispecies in sera of patients with a coexistence of HBsAg and anti-HBs antibodies. Sci. Rep. 2017, 7, 661. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Qian, F.; Yuan, Q.; Li, X.; Wu, W.; Guo, X.; Li, L. Mutations in hepatitis B virus DNA from patients with coexisting HBsAg and anti-HBs. J. Clin. Virol. 2011, 52, 198–203. [Google Scholar] [CrossRef]
- Wu, I.C.; Liu, W.-C.; Chang, T.-T. Applications of next-generation sequencing analysis for the detection of hepatocellular carcinoma-associated hepatitis B virus mutations. J. Biomed. Sci. 2018, 25, 51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.-Y.; Zeng, Z.; Xu, X.-Y.; Zhang, N.-L.; Yu, M.; Gong, W.-B. Mutations in surface and polymerase gene of chronic hepatitis B patients with coexisting HBsAg and anti-HBs. World J. Gastroenterol. 2006, 12, 4219–4223. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, H.; Ning, X.; Gao, F. Sequence analysis of the S gene region in HBV DNA from patients positive for both HBsAg and HBsAb tests. Hepatol. Res. Off. J. Jpn. Soc. Hepatol. 2010, 40, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Nainan, O.V.; Jia, Z. Characterization and distribution of hepatitis B virus genotypes and subtypes in 4 provinces of China. Zhonghua Liu Xing Bing Xue Za Zhi 2001, 22, 348–351. [Google Scholar] [PubMed]
- Zhang, J.-M.; Xu, Y.; Wang, X.-Y.; Yin, Y.-K.; Wu, X.-H.; Weng, X.-H.; Lu, M. Coexistence of hepatitis B surface antigen (HBsAg) and heterologous subtype-specific antibodies to HBsAg among patients with chronic hepatitis B virus infection. Clin. Infect. Dis. 2007, 44, 1161–1169. [Google Scholar] [CrossRef]
- Tabor, E.; Gerety, R.J.; Smallwood, L.A.; Barker, L.F. Coincident hepatitis B surface antigen and antibodies of different subtypes in human serum. J. Immunol. 1977, 118, 369–370. [Google Scholar]
- Wang, S.; Wang, J.; Fan, M.J.; Li, T.Y.; Pan, H.; Wang, X.; Liu, H.K.; Lin, Q.F.; Zhang, J.G.; Guan, L.P.; et al. Identified OAS3 gene variants associated with coexistence of HBsAg and anti-HBs in chronic HBV infection. J. Viral. Hepat. 2018, 25, 904–910. [Google Scholar] [CrossRef]
- Li, Z.; Woo, C.J.; Iglesias-Ussel, M.D.; Ronai, D.; Scharff, M.D. The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev. 2004, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Turner, S.J.; La Gruta, N.L.; Kedzierska, K.; Thomas, P.G.; Doherty, P.C. Functional implications of T cell receptor diversity. Curr. Opin. Immunol. 2009, 21, 286–290. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Q.; Xu, J.H.; Yu, Y.Y.; Lo KK, E.; Felicianna; El-Nezami, H.; Zeng, Z. Human immune repertoire in hepatitis B virus infection. World J. Gastroenterol. 2021, 27, 3790–3801. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-Q.; Zhang, X.-X. Research progress in host immune response during hepatitis B virus infection. Chin. Med. J. 2020, 133, 2746–2748. [Google Scholar] [CrossRef] [PubMed]
- Valkiers, S.; Van Houcke, M.; Laukens, K.; Meysman, P. ClusTCR: A Python interface for rapid clustering of large sets of CDR3 sequences with unknown antigen specificity. Bioinformatics 2021, btab446. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, Z.; Li, D.; Wang, A.; Su, H.; Shao, Z.; Zhang, J.; Ji, Z.; Gao, J.; Choi, B.C.K.; Yan, Y. Epidemiological characteristics of the carriers with coexistence of HBsAg and anti-HBs based on a community cohort study. J. Viral. Hepat. 2016, 23, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Yu, H.-G.; Li, Y.-X.; Cui, N.; Dai, J.-F.; Yu, J.-P. Sequence analysis of the HBV S protein in Chinese patients with coexisting HBsAg and anti-HBs antibodies. J. Med. Virol. 2015, 87, 2067–2073. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Chen, W.; Xu, C.; Yu, X.; Xi, Y. Virology and serological characteristics of chronic Hepatitis B patients with the co-existence of HBsAg and anti-HBs antibodies. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Heijtink, R.A.; van Hattum, J.; Schalm, S.W.; Masurel, N. Co-occurrence of HBsAg and anti-HBs: Two consecutive infections or a sign of advanced chronic liver disease? J. Med. Virol. 1982, 10, 83–90. [Google Scholar] [CrossRef]
- Lee, S.-A.; Kim, K.; Kim, H.; Kim, B.-J. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J. Hepatol. 2012, 56, 63–69. [Google Scholar] [CrossRef]
- Shields, P.L.; Owsianka, A.; Carman, W.F.; Boxall, E.; Hubscher, S.G.; Shaw, J.; O’Donnell, K.; Elias, E.; Mutimer, D.J. Selection of hepatitis B surface “escape” mutants during passive immune prophylaxis following liver transplantation: Potential impact of genetic changes on polymerase protein function. Gut 1999, 45, 306–309. [Google Scholar] [CrossRef]
- Domingo, E.; Sheldon, J.; Perales, C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Wang, M.-J.; Yang, Z.-T.; Yu, D.-M.; Han, Y.; Huang, D.; Zhang, D.-H.; Zhang, X.-X. Clinical features and viral quasispecies characteristics associated with infection by the hepatitis B virus G145R immune escape mutant. Emerg. Microbes Infect. 2017, 6, e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Li, Y.; Ye, J.; Wang, J.; Lu, H.; Yao, X. Characterization of the TCR β Chain Repertoire in Peripheral Blood from Hepatitis B Vaccine Responders and Non-Responders. J. Inflamm. Res. 2022, 15, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Maru, Y.; Yokosuka, O.; Imazeki, F.; Saisho, H.; Omata, M. Analysis of T cell receptor variable regions and complementarity determining region 3 of infiltrating T lymphocytes in the liver of patients with chronic type B hepatitis. Intervirology 2003, 46, 277–288. [Google Scholar] [CrossRef]
- Yousfi Monod, M.; Giudicelli, V.; Chaume, D.; Lefranc, M.-P. IMGT/JunctionAnalysis: The first tool for the analysis of the immunoglobulin and T cell receptor complex V–J and V–D–J junctions. Bioinformatics 2004, 20 (Suppl. S1), i379–i385. [Google Scholar] [CrossRef] [Green Version]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Sherwood, A.M.; Emerson, R.O.; Scherer, D.; Habermann, N.; Buck, K.; Staffa, J.; Desmarais, C.; Halama, N.; Jaeger, D.; Schirmacher, P.; et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol. Immunother. 2013, 62, 1453–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]

| Group | DP (26) | SP (25) | CHB (25) | p-Value |
|---|---|---|---|---|
| Gender, male, % (n) | 57.69 (15) | 52.00 (13) | 60.00 (15) | 0.841 |
| Age, years | 38.38 ± 11.82 | 40.28 ± 8.01 | 40.68 ± 8.51 | 0.278 |
| HBeAg positive ratio, % (n) | 73.08 (19) | - | 40.00 (10) | 0.017 |
| HBeAg (COI) | 14.69 (0.48–1150.25) | - | 0.40 (0.22–302.88) | 0.121 |
| HBsAg (IU/mL) | 2578.00 (1186.00–6989.75) | - | 250.00 (250.00–1731.00) | 0.001 |
| Anti-HBs (IU/mL) | 100.92 (35.11–249.18) | 125.02 (25.05–401.83) | - | 0.474 |
| HBV DNA (log10 IU/mL) | 4.51 ± 2.06 | - | 4.12 ± 2.14 | 0.466 |
| Group | Sequence | Width | Sites | E-Value |
|---|---|---|---|---|
| DP | SSGGGGNEQ | 9 | 347,562 | 1.7 × 10−1963 |
| CASSLG | 6 | 75,967 | 3.2 × 10−206 | |
| NYGYTF | 6 | 21,764 | 4.8 × 10−063 | |
| NSPLHF | 6 | 4350 | 8.6 × 10−008 | |
| NTEAFF | 6 | 3965 | 1.1 × 10+001 | |
| SP | SSGGGGNEQ | 9 | 499,849 | 1.2 × 10−1887 |
| CASSLG | 6 | 51,033 | 2.0 × 10−083 | |
| ANYGYT | 6 | 379 | 1.1 × 10+006 | |
| YYGYTF | 6 | 166 | 2.4 × 10+006 | |
| GAGPLT | 6 | 89 | 2.4 × 10+006 | |
| CHB | SSGGGGNE | 8 | 485,120 | 1.5 × 10−1783 |
| CASSLG | 6 | 58,698 | 1.3 × 10−064 | |
| NSPLHF | 6 | 4194 | 1.8 × 10−024 | |
| SYEQYF | 6 | 12,060 | 1.8 × 10−017 | |
| NYGYTF | 6 | 12,270 | 2.7 × 10−013 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhan, Q.; Chang, L.; Wu, J.; Zhang, Z.; Xu, J.; Yu, Y.; Feng, Z.; Zeng, Z. T-Cell Receptor β Chain and B-Cell Receptor Repertoires in Chronic Hepatitis B Patients with Coexisting HBsAg and Anti-HBs. Pathogens 2022, 11, 727. https://doi.org/10.3390/pathogens11070727
Zhan Q, Chang L, Wu J, Zhang Z, Xu J, Yu Y, Feng Z, Zeng Z. T-Cell Receptor β Chain and B-Cell Receptor Repertoires in Chronic Hepatitis B Patients with Coexisting HBsAg and Anti-HBs. Pathogens. 2022; 11(7):727. https://doi.org/10.3390/pathogens11070727
Chicago/Turabian StyleZhan, Qiao, Le Chang, Jian Wu, Zhiyuan Zhang, Jinghang Xu, Yanyan Yu, Zhenru Feng, and Zheng Zeng. 2022. "T-Cell Receptor β Chain and B-Cell Receptor Repertoires in Chronic Hepatitis B Patients with Coexisting HBsAg and Anti-HBs" Pathogens 11, no. 7: 727. https://doi.org/10.3390/pathogens11070727
APA StyleZhan, Q., Chang, L., Wu, J., Zhang, Z., Xu, J., Yu, Y., Feng, Z., & Zeng, Z. (2022). T-Cell Receptor β Chain and B-Cell Receptor Repertoires in Chronic Hepatitis B Patients with Coexisting HBsAg and Anti-HBs. Pathogens, 11(7), 727. https://doi.org/10.3390/pathogens11070727







