Characterization of Vaginal Microbiota in Third Trimester Premature Rupture of Membranes Patients through 16S rDNA Sequencing
Abstract
:1. Introduction
2. Results
2.1. Demographical and Clinical Data
2.2. 16S rDNA V3–V4 Gene Sequencing Statistics
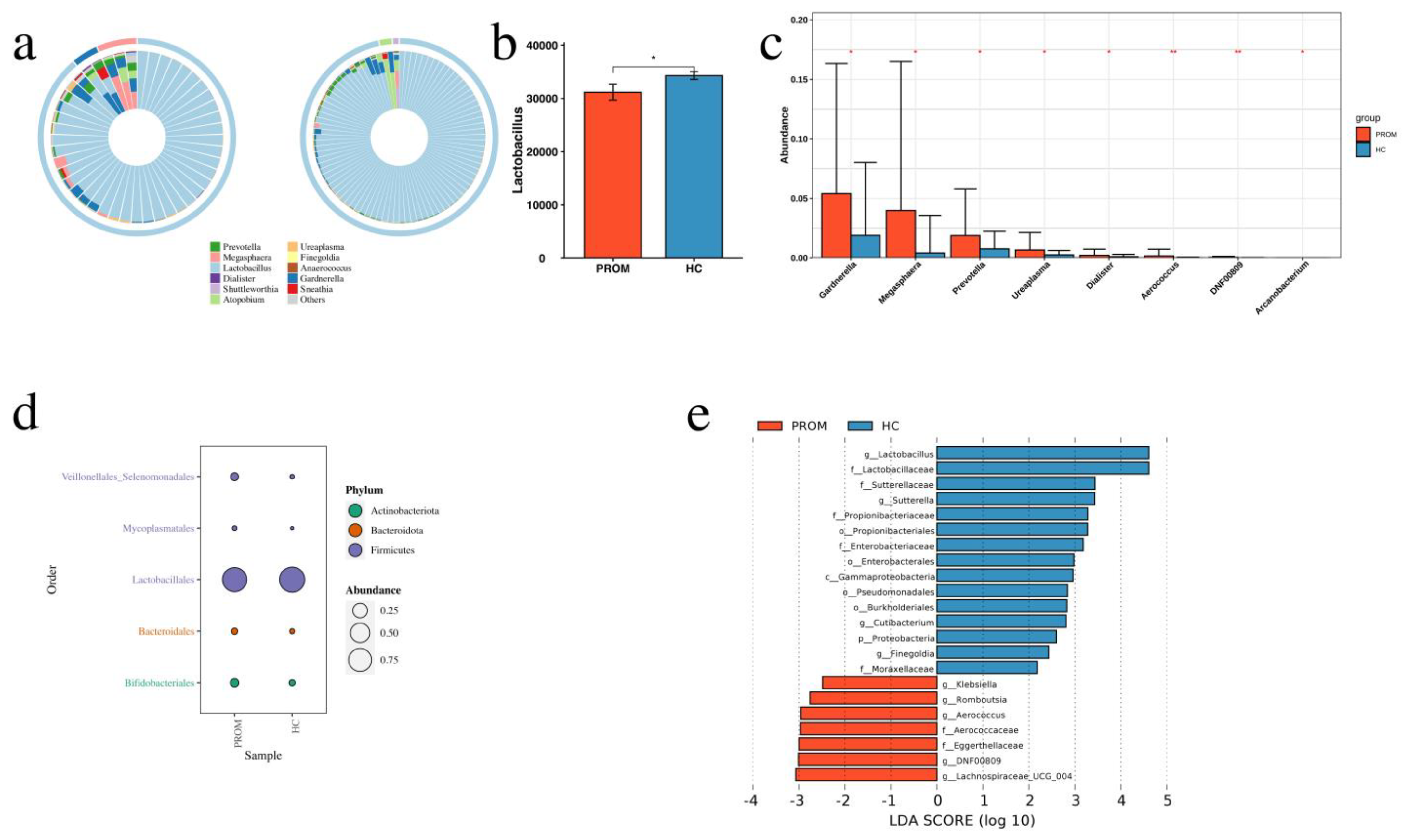
2.3. Different Microbiome Composition between PROM and HC Groups
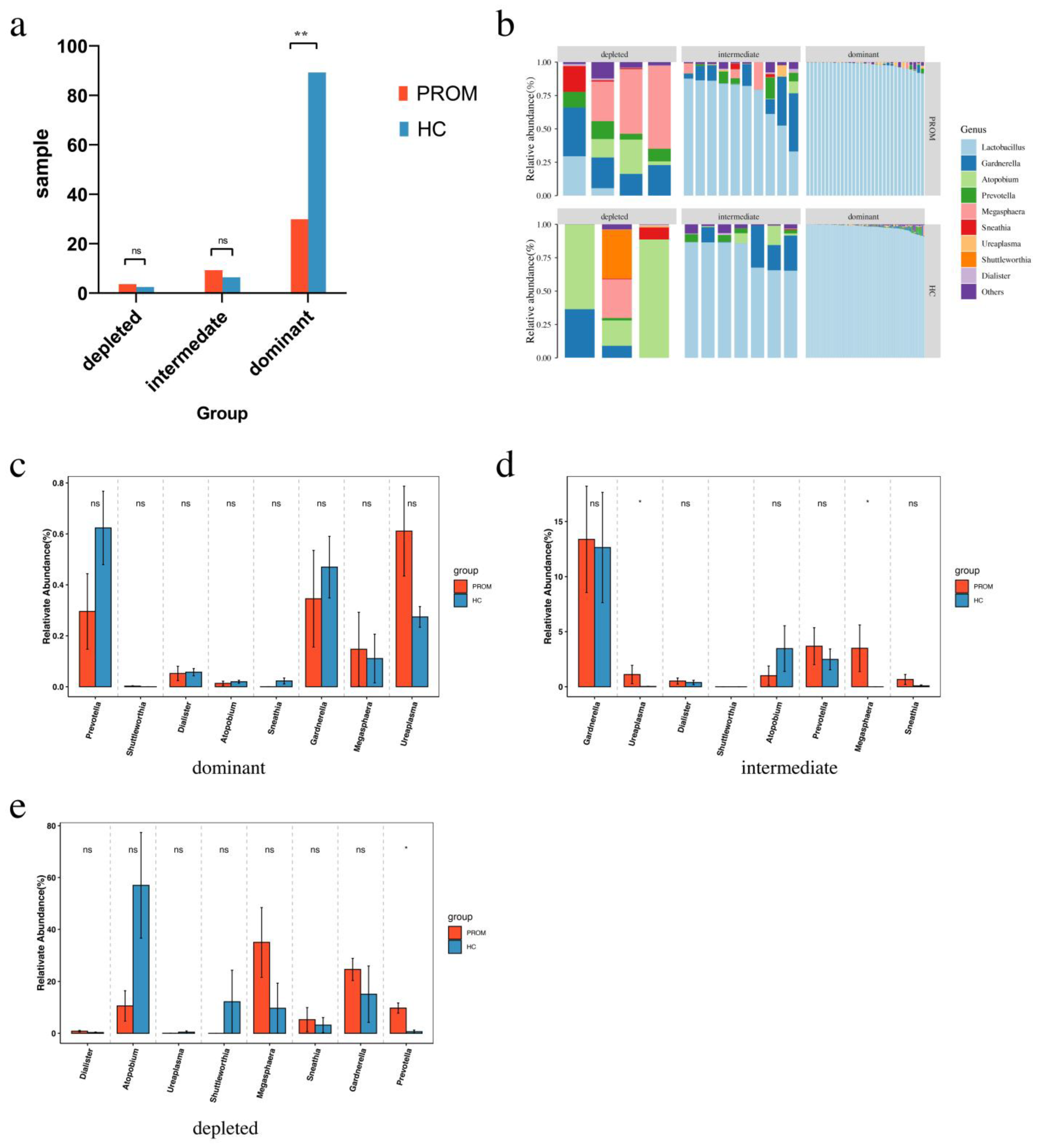
2.4. Bioinformatics Analysis among Groups with Different Abundance of Lactobacillus
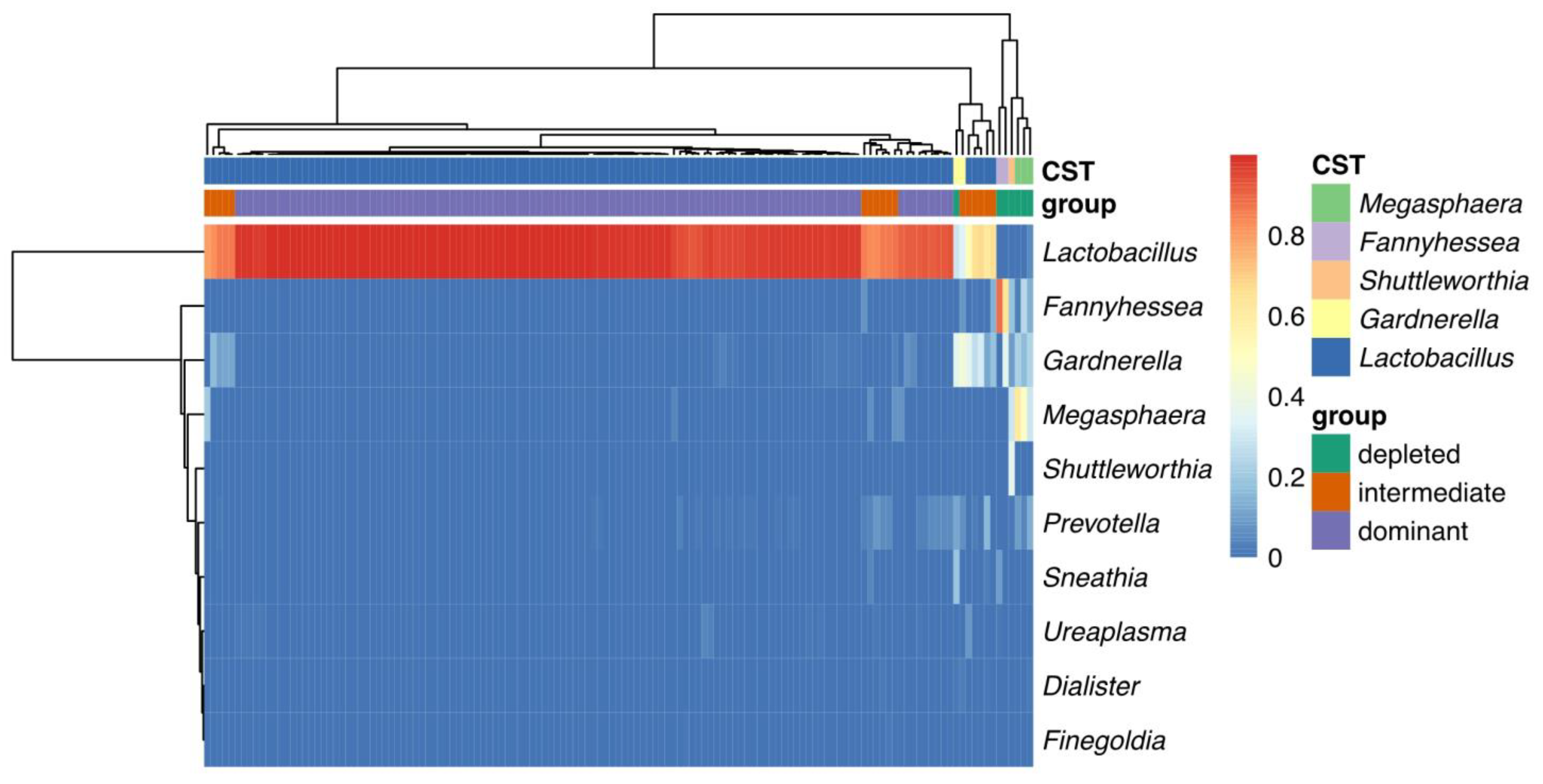
2.5. Cross-Analysis between the Two Grouping Methods
2.6. Intergrated Analysis of Vaginal Microbiome in All Samples
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Prenatal Examination and Delivery
4.3. PROM Cases and Healthy Controls Selection
4.4. DNA Extraction
4.5. 16S rDNA Gene Sequencing and Data Processing
4.6. Taxonomic Assignment
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ehsanipoor, R. Practice Bulletin No. 172: Premature Rupture of Membranes. Obstet. Gynecol. 2016, 128, e165–e177. [Google Scholar] [CrossRef]
- Genovese, C.; Corsello, S.; Nicolosi, D.; Aidala, V.; Falcidia, E.; Tempera, G. Alterations of the vaginal microbiota in the third trimester of pregnancy and pPROM. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3336–3343. [Google Scholar] [PubMed]
- Mendz, G.L.; Kaakoush, N.O.; Quinlivan, J.A. Bacterial aetiological agents of intra-amniotic infections and preterm birth in pregnant women. Front. Cell Infect. Microbiol. 2013, 3, 58. [Google Scholar] [CrossRef] [Green Version]
- Chandiramani, M.; Bennett, P.R.; Brown, R.; Lee, Y.S.; Macintyre, D.A. Vaginal Microbiome–Pregnant Host Interactions Determine a Significant Proportion of Preterm Labour. Fetal Matern. Med. Rev. 2014, 25, 73–78. [Google Scholar] [CrossRef]
- Lahiri, S.S. Bacterial toxins—An overview. J. Nat. Toxins 2000, 9, 381–408. [Google Scholar] [PubMed]
- Holmstrom, E.; Myntti, T.; Sorsa, T.; Kruit, H.; Juhila, J.; Paavonen, J.; Rahkonen, L.; Stefanovic, V. Cervical and Amniotic Fluid Matrix Metalloproteinase-8 and Interleukin-6 Concentrations in Preterm Pregnancies with or without Preterm Premature Rupture of Membranes. Fetal Diagn. Ther. 2019, 46, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Seta, F.; Parazzini, F.; De Leo, R.; Banco, R.; Maso, G.P.; De Santo, D.; Sartore, A.; Stabile, G.; Inglese, S.; Tonon, M.; et al. Lactobacillus plantarum P17630 for preventing Candida vaginitis recurrence: A retrospective comparative study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014, 182, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Kitaya, K.; Nagai, Y.; Arai, W.; Sakuraba, Y.; Ishikawa, T. Characterization of Microbiota in Endometrial Fluid and Vaginal Secretions in Infertile Women with Repeated Implantation Failure. Mediat. Inflamm. 2019, 2019, 4893437. [Google Scholar] [CrossRef] [Green Version]
- Parry, S.; Strauss, J.F., 3rd. Premature rupture of the fetal membranes. N. Engl. J. Med. 1998, 338, 663–670. [Google Scholar] [CrossRef]
- Fettweis, J.M.; Serrano, M.G.; Brooks, J.P.; Edwards, D.J.; Girerd, P.H.; Parikh, H.I.; Huang, B.; Arodz, T.J.; Edupuganti, L.; Glascock, A.L.; et al. The vaginal microbiome and preterm birth. Nat. Med. 2019, 25, 1012–1021. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, S.; Pryor, K.; Antczak, B.; Truong, T.; Murtha, A.; Seed, P. The relationship of cervical microbiota diversity with race and disparities in preterm birth. J. Neonatal Perinatal Med. 2018, 11, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, J.W.; Xia, H.W.; Zhang, H.X.; Betran, A.P.; Zhang, L.; Hua, X.L.; Feng, L.P.; Chen, D.; Sun, K.; et al. Preterm Birth in China Between 2015 and 2016. Am. J. Public Health 2019, 109, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, X.; Li, X.; Liang, H.; Xu, H. The clinical management and outcome of term premature rupture of membrane in East China: Results from a retrospective multicenter study. Int. J. Clin. Exp. Med. 2015, 8, 6212–6217. [Google Scholar] [PubMed]
- Jiang, H.; Lu, C.; Zhou, J.; Zhang, W. Cesarean section and pregnancy outcomes of preterm premature rupture of membranes under different fertility policies in China. Transl. Pediatr. 2021, 10, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, N.S.; Callaghan, W.; Johnson, C.; Williams, L. Racial, ethnic, and economic disparities in the prevalence of pregnancy complications. Matern. Child. Health J. 2009, 13, 198–205. [Google Scholar] [CrossRef]
- Ma, Y.; Lan, G.; Li, C.; Cambaza, E.M.; Liu, D.; Ye, X.; Chen, S.; Ding, T. Stress tolerance of Staphylococcus aureus with different antibiotic resistance profiles. Microb. Pathog. 2019, 133, 103549. [Google Scholar] [CrossRef]
- Brown, R.G.; Al-Memar, M.; Marchesi, J.R.; Lee, Y.S.; Smith, A.; Chan, D.; Lewis, H.; Kindinger, L.; Terzidou, V.; Bourne, T.; et al. Establishment of vaginal microbiota composition in early pregnancy and its association with subsequent preterm prelabor rupture of the fetal membranes. Transl. Res. 2019, 207, 30–43. [Google Scholar] [CrossRef] [Green Version]
- Kindinger, L.M.; MacIntyre, D.A.; Lee, Y.S.; Marchesi, J.R.; Smith, A.; McDonald, J.A.; Terzidou, V.; Cook, J.R.; Lees, C.; Israfil-Bayli, F.; et al. Relationship between vaginal microbial dysbiosis, inflammation, and pregnancy outcomes in cervical cerclage. Sci. Transl. Med. 2016, 8, 350ra102. [Google Scholar] [CrossRef] [Green Version]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4680–4688. [Google Scholar] [CrossRef] [Green Version]
- Romero, R.; Hassan, S.S.; Gajer, P.; Tarca, A.L.; Fadrosh, D.W.; Bieda, J.; Chaemsaithong, P.; Miranda, J.; Chaiworapongsa, T.; Ravel, J. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome 2014, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Biegert, G.; Karpinets, T.; Wu, X.; Alam, M.B.E.; Sims, T.T.; Yoshida-Court, K.; Lynn, E.J.; Yue, J.; Medrano, A.D.; Petrosino, J.; et al. Diversity and composition of gut microbiome of cervical cancer patients by 16S rRNA and whole-metagenome sequencing. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kimberlin, D.F.; Andrews, W.W. Bacterial vaginosis: Association with adverse pregnancy outcome. Semin. Perinatol. 1998, 22, 242–250. [Google Scholar] [CrossRef]
- Tabatabaei, N.; Eren, A.M.; Barreiro, L.B.; Yotova, V.; Dumaine, A.; Allard, C.; Fraser, W.D. Vaginal microbiome in early pregnancy and subsequent risk of spontaneous preterm birth: A case-control study. BJOG 2019, 126, 349–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paramel Jayaprakash, T.; Wagner, E.C.; van Schalkwyk, J.; Albert, A.Y.; Hill, J.E.; Money, D.M. High Diversity and Variability in the Vaginal Microbiome in Women following Preterm Premature Rupture of Membranes (PPROM): A Prospective Cohort Study. PLoS ONE 2016, 11, e0166794. [Google Scholar] [CrossRef] [PubMed]
- Ren, D.; Li, C.; Qin, Y.; Yin, R.; Du, S.; Liu, H.; Zhang, Y.; Wang, C.; Rong, F.; Jin, N. Evaluation of immunomodulatory activity of two potential probiotic Lactobacillus strains by in vivo tests. Anaerobe 2015, 35, 22–27. [Google Scholar] [CrossRef]
- Sillanpää, J.; Martínez, B.; Antikainen, J.; Toba, T.; Kalkkinen, N.; Tankka, S.; Lounatmaa, K.; Keränen, J.; Höök, M.; Westerlund-Wikström, B.; et al. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 2000, 182, 6440–6450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendling, W. Vaginal Microbiota. Adv. Exp. Med. Biol. 2016, 902, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Kim, M.H.; Lee, W.I.; Kang, S.Y.; Jeon, Y.L. Prevalence and Antibiotic Susceptibility of Mycoplasma hominis and Ureaplasma urealyticum in Pregnant Women. Yonsei Med. J. 2016, 57, 1271–1275. [Google Scholar] [CrossRef] [PubMed]
- Sprong, K.E.; Mabenge, M.; Wright, C.A.; Govender, S. Ureaplasma species and preterm birth: Current perspectives. Crit. Rev. Microbiol. 2020, 46, 169–181. [Google Scholar] [CrossRef] [PubMed]
- King, A.E.; Critchley, H.O.; Kelly, R.W. Innate immune defences in the human endometrium. Reprod. Biol. Endocrinol. 2003, 1, 116. [Google Scholar] [CrossRef] [Green Version]
- King, A.E.; Wheelhouse, N.; Cameron, S.; McDonald, S.E.; Lee, K.F.; Entrican, G.; Critchley, H.O.; Horne, A.W. Expression of secretory leukocyte protease inhibitor and elafin in human fallopian tube and in an in-vitro model of Chlamydia trachomatis infection. Hum. Reprod. 2009, 24, 679–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, N.; Guo, R.; Yao, Y.; Jin, M.; Cheng, Y.; Ling, Z. Lactobacillus iners Is Associated with Vaginal Dysbiosis in Healthy Pregnant Women: A Preliminary Study. Biomed. Res. Int. 2019, 2019, 6079734. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.A.; Walther-Antonio, M.; MacLean, A.M.; Gohl, D.M.; Beckman, K.B.; Chen, J.; White, B.; Creedon, D.J.; Chia, N. Persistent microbial dysbiosis in preterm premature rupture of membranes from onset until delivery. PeerJ 2015, 3, e1398. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in Low Microbial Biomass Microbiome Studies: Issues and Recommendations. Trends Microbiol. 2019, 27, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Bang, J.Y.; Park, G.W.; Choi, D.S.; Kang, J.S.; Kim, H.J.; Park, K.S.; Lee, J.O.; Kim, Y.K.; Kwon, K.H.; et al. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 2007, 7, 3143–3153. [Google Scholar] [CrossRef]
- Sewe, S.O.; Silva, G.; Sicat, P.; Seal, S.E.; Visendi, P. Trimming and Validation of Illumina Short Reads Using Trimmomatic, Trinity Assembly, and Assessment of RNA-Seq Data. Methods Mol. Biol. 2022, 2443, 211–232. [Google Scholar] [CrossRef]
- Sharma, R.; Kumar, A.; Singh, N.; Sharma, K. 16S rRNA gene profiling of rhizospheric microbial community of Eichhornia crassipes. Mol. Biol. Rep. 2021, 48, 4055–4064. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Majaneva, M.; Hyytiäinen, K.; Varvio, S.L.; Nagai, S.; Blomster, J. Bioinformatic Amplicon Read Processing Strategies Strongly Affect Eukaryotic Diversity and the Taxonomic Composition of Communities. PLoS ONE 2015, 10, e0130035. [Google Scholar] [CrossRef]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characteristics | Whole Study Cohort (n = 441) | 16S rDNA Amplicon Sequencing (n = 135) | ||||
|---|---|---|---|---|---|---|
| PROM Cases (n = 84, 19.05%) | Controls (n = 342, 77.55%) | Lost to Follow-Up (n = 15, 3.40%) | PROM Cases (n = 45) | Controls (n = 90) | p-Value | |
| Maternal age a | 29.06 ± 4.08 | 29.91 ± 4.41 | 29.67 ± 4.3 | 28.96 ± 3.80 | 29.70 ± 3.66 | 0.251 † |
| Nulliparity | 55 (65.48%) | 180 (52.63%) | 10 (66.67%) | 32 (71.11%) | 49 (54.44%) | 0.09 |
| White blood cell at admission (×109/L) | 9.82 ± 2.8 | 9.4 ± 2.07 | 9.4 ± 1.65 | 10.01 ± 3.00 | 9.49 ± 2.67 | 0.242 † |
| Positive genital cultures at admission | 24(28.57%) | 57(16.67%) | 4 (26.67%) | 7 (15.56%) | 12 (13.33%) | 0.795 |
| Gestational age at sampling (weeks) a | 32.84 ± 1.53 | 32.54 ± 1.43 | 32.81 ± 1.47 | 33.18 ± 1.7 | 33.01 ± 1.59 | 0.553 † |
| Steroid administration | 6 (7.14%) | 5 (1.46%) | - | 0 | 0 | - |
| Tocolysis treatment | 17 (20.24%) | 61 (17.83%) | - | 8 (17.78%) | 7 (7.78%) | 0.099 ‡ |
| Gestational age at delivery (weeks) a | 38.46 ± 1.60 | 38.93 ± 1.05 | - | 39.27 ± 1.45 | 39.65 ± 0.97 | 0.098 † |
| Latency from sampling to delivery (weeks) a | 5.16 ± 6.23 | 6.08 ± 2.00 | - | 5.84 ± 2.36 | 6.39 ± 0.14 | 0.148 † |
| BMI in the first trimester(kg/m2) a | 22.15 ± 2.90 | 22.23 ± 4.98 | - | 22.19 ± 3.31 | 21.74 ± 2.73 | 0.140 † |
| BMI in third trimester (kg/m2) a | 26.36 ± 6.33 | 26.89 ± 5.16 | - | 27.30 ± 3.84 | 27.66 ± 2.98 | 0.537 † |
| Estriol at sampling a | 7.22 ± 0.44 | 7.61 ± 1.66 | - | 7.22 ± 2.79 | 7.38 ± 2.78 | 0.748 † |
| Number of deliveries a | 1.29 ± 0.17 | 1.44 ± 0.54 | - | 1.31 ± 0.51 | 1.47 ± 0.56 | 0.101 † |
| spontaneous abortion a | 0.91 ± 0.05 | 0.67 ± 0.73 | - | 1.00 ± 0.16 | 0.66 ± 0.81 | 0.053 † |
| Baby weight at birth (g) a | 3318.78 ± 525.96 | 3397.95 ± 441.45 | - | 3031 ± 368.75 | 3041 ± 379.09 | 0.168 † |
| Baby gender (male/female) | 48/36 (57.14%/42.86%) | 226/106 (66.08%/30.99%) | - | 25/20 (55.56%/44.44%) | 48/42 (53.33%/46.67%) | 0.84 ‡ |
| Spontaneous abortion (number) a | 0.93 ± 1.12 | 0.67 ± 0.73 | 1.00 ± 1.15 | 0.66 ± 0.91 | 0.053 † | |
| gestational diabetes | 15 (17.86%) | 41 (11.99%) | - | 0 | 0 | - |
| preeclampsia | 16 (19.05%) | 38 (11.11%) | 0 | 0 | - | |
| Estimated Coeffieicent (95% CI) | Adjusted p-Value | |
|---|---|---|
| Lactobacillus | −0.09 (−0.06 to −0.01) | 0.04 |
| Fannyhessea | −0.01 (−0.05 to 0.03) | 0.57 |
| Gardnerella | 0.04 (0.01 to 0.06) | 0.02 |
| Megasphaera | 0.04 (0.01 to 0.06) | 0.01 |
| Shuttleworthia | −0.004 (−0.02 to 0.007) | 0.48 |
| Prevotella | 0.11 (0.002 to 0.02) | 0.02 |
| Sneathia | 0.005 (−0.002 to 0.01) | 0.16 |
| Ureaplasma | 0.004 (0.001 to 0.007) | 0.01 |
| Dialister | 0.001 (0.001 to 0.0003) | 0.04 |
| Finegoldia | −0.47 × 10−5 (−0.0009 to 0.0008) | 0.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Chen, J.; Chen, Y.; Jiang, S.; Xu, H.; Zhan, H.; Ren, Y.; Xu, D.; Xu, Z.; Chen, D. Characterization of Vaginal Microbiota in Third Trimester Premature Rupture of Membranes Patients through 16S rDNA Sequencing. Pathogens 2022, 11, 847. https://doi.org/10.3390/pathogens11080847
Liu L, Chen J, Chen Y, Jiang S, Xu H, Zhan H, Ren Y, Xu D, Xu Z, Chen D. Characterization of Vaginal Microbiota in Third Trimester Premature Rupture of Membranes Patients through 16S rDNA Sequencing. Pathogens. 2022; 11(8):847. https://doi.org/10.3390/pathogens11080847
Chicago/Turabian StyleLiu, Lou, Jiale Chen, Yu Chen, Shiwen Jiang, Hanjie Xu, Huiying Zhan, Yongwei Ren, Dexiang Xu, Zhengfeng Xu, and Daozhen Chen. 2022. "Characterization of Vaginal Microbiota in Third Trimester Premature Rupture of Membranes Patients through 16S rDNA Sequencing" Pathogens 11, no. 8: 847. https://doi.org/10.3390/pathogens11080847
APA StyleLiu, L., Chen, J., Chen, Y., Jiang, S., Xu, H., Zhan, H., Ren, Y., Xu, D., Xu, Z., & Chen, D. (2022). Characterization of Vaginal Microbiota in Third Trimester Premature Rupture of Membranes Patients through 16S rDNA Sequencing. Pathogens, 11(8), 847. https://doi.org/10.3390/pathogens11080847






