Occurrence and Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Strains Isolated from Hospitalized Patients in Poland—A Single Centre Study
Abstract
:1. Introduction
2. Results
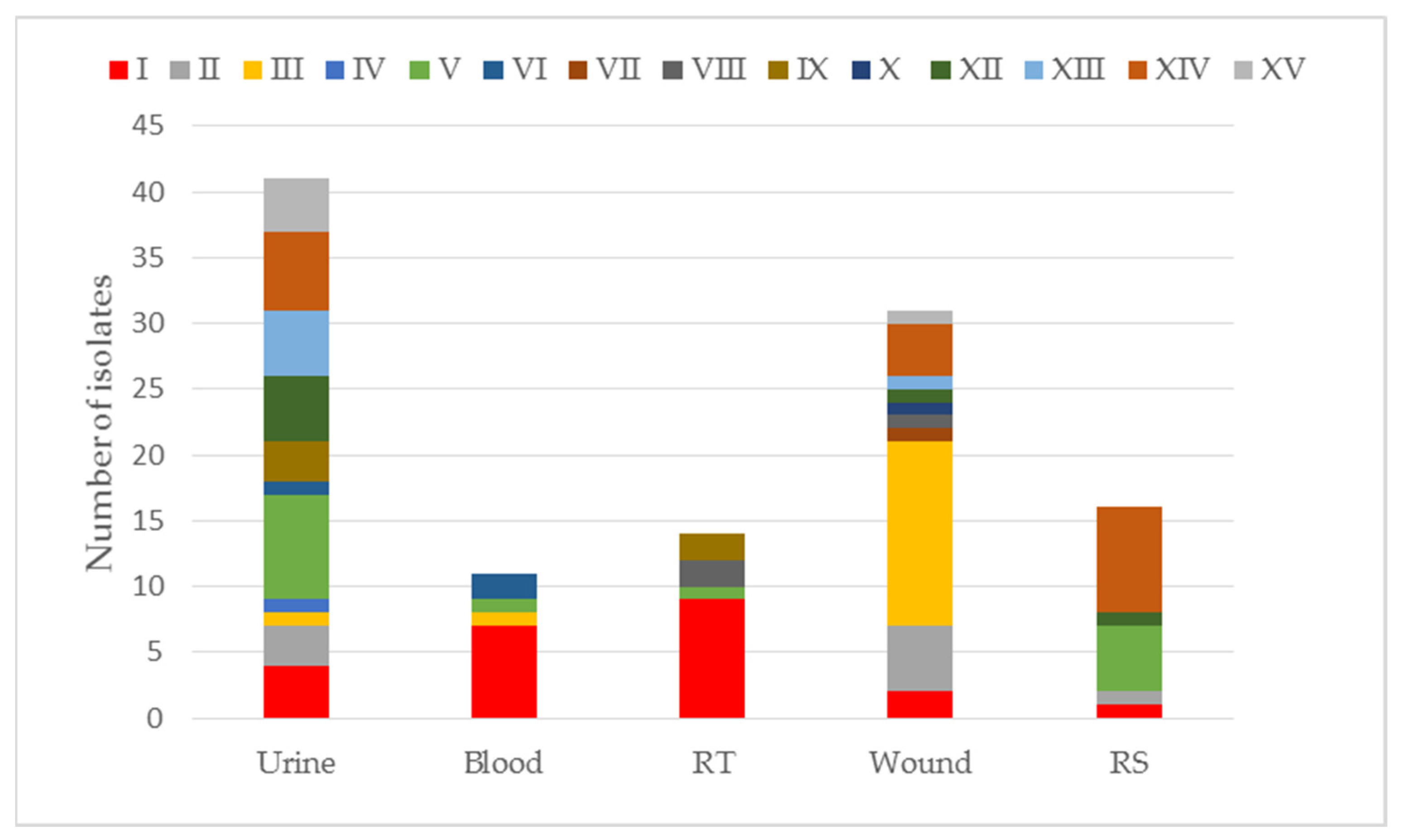
2.1. Assessment of the Antibiotic Susceptibility of K. pneumoniae
2.2. Susceptibility Assessment for Ertapenem
2.3. Detection of Carbapenemases
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Microbiological Assays
4.2.1. Automated System
4.2.2. Disc Diffusion Method
4.2.3. Detection of Carbapenemases
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef]
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.M.; Silva, B.N.M.D.; Barbosa, G.; Barreiro, E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 208, 112–120. [Google Scholar] [CrossRef]
- Baraniak, A.; Izdebski, R.; Fiett, J.; Gawryszewska, I.; Bojarska, K.; Herda, M.; Literacka, E.; Żabicka, D.; Tomczak, H.; Pewińska, N.; et al. NDM-producing Enterobacteriaceae in Poland, 2012–2014: Inter-regional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J. Antimicrob. Chemother. 2016, 71, 85–91. [Google Scholar] [CrossRef] [PubMed]
- El-Gamal, M.I.; Brahim, I.; Hisham, N.; Aladdin, R.; Mohammed, H.; Bahaaeldin, A. Recent updates of carbapenem antibiotics. Eur. J. Med. Chem. 2017, 131, 185–195. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 13, S521–S528. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 21, 1. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 2010, 300, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Abbas, H.A.; Kadry, A.A.; Shaker, G.H.; Goda, R.M. Impact of specific inhibitors on metallo-β-carbapenemases detected in Escherichia coli and Klebsiella pneumoniae isolates. Microb. Pathog. 2019, 132, 266–274. [Google Scholar] [CrossRef]
- Logan, L.K.; Weinstein, R.A. The epidemiology of carbapenem-resistant enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Stoesser, N.; Sheppard, A.E.; Peirano, G.; Anson, L.W.; Pankhurst, L.; Sebra, R.; Phan, H.T.T.; Kasarskis, A.; Mathers, A.J.; Peto, T.E.A.; et al. Genomic epidemiology of global Klebsiella pneumoniae carbapenemase (KPC)-producing Escherichia coli. Sci. Rep. 2017, 7, 5917. [Google Scholar] [CrossRef] [PubMed]
- Elshamy, A.A.; Aboshanab, K.M. A review on bacterial resistance to carbapenems: Epidemiology, detection and treatment options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Criteria for Diagnosis of an Outbreak in a Hospital/Health Care Facility Caused by Carbapenamase-Producing Strains of Enterobacteriaceae (CPE). Guidelines of the National Antibiotic Protection Program. Available online: http://antybiotyki.edu.pl/wp-content/uploads/dokumenty/CPE_NDM-Ognisko-epidemiczne.pdf (accessed on 31 May 2021).
- ECDC. Antimicrobial consumption in the EU/EEA—Annual Epidemiological Report 2019. Stockholm 2020, 11, 1–25. [Google Scholar]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Fiett, J.; Baraniak, A.; Izdebski, R.; Sitkiewicz, I.; Żabicka, D.; Meler, A.; Filczak, K.; Hryniewicz, W.; Gniadkowski, M. The first NDM metallo-β-lactamase-producing Enterobacteriaceae isolate in Poland: Evolution of IncFII-type plasmids carrying the bla(NDM-1) gene. Antimicrob. Agents Chemother. 2014, 58, 1203–1207. [Google Scholar] [CrossRef]
- Baraniak, A.; Grabowska, A.; Izdebski, R.; Fiett, J.; Herda, M.; Bojarska, K.; Żabicka, D.; Kania-Pudło, M.; Młynarczyk, G.; Żak-Puławska, Z.; et al. KPC-PL Study Group. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008–2009. Antimicrob. Agents Chemother. 2011, 55, 5493–5499. [Google Scholar] [CrossRef]
- Cantón, R.; Akóva, M.; Carmeli, Y.; Giske, C.G.; Glupczynski, Y.; Gniadkowski, M.; Livermore, D.M.; Miriagou, V.; Naas, T.; Rossolini, G.M.; et al. European Network on Carbapenemases. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2012, 18, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Baraniak, A.; Izdebski, R.; Zabicka, D.; Bojarska, K.; Górska, S.; Literacka, E.; Fiett, J.; Hryniewicz, W.; Gniadkowski, M. KPC-PL2 Study Group. Multiregional dissemination of KPC-producing Klebsiella pneumoniae ST258/ST512 genotypes in Poland, 2010–2014. J. Antimicrob. Chemother. 2017, 72, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Van Duin, D.; Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Literacka, E.; Żabicka, D.; Hryniewicz, W.; Gniadkowski, M. Report of the National Reference Center for Antimicrobial Susceptibility. Data from the National Reference Center for Antimicrobial Susceptibility on Enterobacterales, Producing NDM, KPC, VIM and OXA-48 Carbapenemases in Poland in 2006–2018. 2019. Available online: https://korld.nil.gov.pl/pdf/Raport%20KORLD%202019_EL_2.pdf (accessed on 30 June 2019).
- Hammoudi Halat, D.; Ayoub Moubareck, C. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef]
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Global Antimicrobial Resistance Surveillance System; WHO: Geneva, Switzerland, 2015; p. 36. [Google Scholar]
- World Health Orgnazation (WHO). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report; WHO: Geneva, Switzerland, 2020; p. 144. [Google Scholar]
- World Health Orgnazation (WHO). Situation Analysis on Antimicrobial Resistance in the South-East Asia Region; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Orgnazation (WHO). Central Asian and Eastern European Surveillance of Antimicrobial Resistance; WHO: Geneva, Switzerland, 2018; p. 39. [Google Scholar]
- Żabicka, D.; Literacka, E.; Gniadkowski, M.; Hryniewicz, W. Report of the National Reference Center for Antimicrobial Susceptibility. Occurrence of Enterobacteriaceae (mainly Klebsiella pneumoniae), producing New Delhi carbapenemase (NDM) in Poland in Q1–Q3 2017. KORLD. 2017. Available online: http://www.korld.edu.pl/pdf/Raport_NDM_18-12-2017 (accessed on 20 December 2017).
- Izdebski, R.; Sitkiewicz, M.; Urbanowicz, P.; Krawczyk, M.; Brisse, S.; Gniadkowski, M. Genomic background of the Klebsiella pneumoniae NDM-1 outbreak in Poland, 2012–2018. J. Antimicrob. Chemother. 2020, 75, 3156–3162. [Google Scholar] [CrossRef] [PubMed]
- Ochońska, D.; Klamińska-Cebula, H.; Dobrut, A.; Bulanda, M.; Brzychczy-Włoch, M. Clonal Dissemination of KPC-2, VIM-1, OXA-48-Producing Klebsiella pneumoniae ST147 in Katowice, Poland. Pol. J. Microbiol. 2021, 70, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Ambretti, S.; Bassetti, M.; Clerici, P.; Petrosillo, N.; Tumietto, F.; Viale, P.; Rossolini, G.M. Screening for carriage of carbapenem-resistant Enterobacteriaceae in settings of high endemicity: A position paper from an Italian working group on CRE infections. Antimicrob Resist. Infect. Control 2019, 8, 136. [Google Scholar] [CrossRef]
- Lin, Q.; Wu, M.; Yu, H.; Jia, X.; Zou, H.; Ma, D.; Niu, S.; Huang, S. Clinical and Microbiological Characterization of Carbapenem-Resistant Enterobacteriales: A Prospective Cohort Study. Front. Pharmacol. 2021, 12, 716324. [Google Scholar] [CrossRef]
- Loqman, S.; Soraa, N.; Diene, S.M.; Rolain, J.-M. Dissemination of Carbapenemases (OXA-48, NDM and VIM) Producing Enterobacteriaceae Isolated from the Mohamed VI University Hospital in Marrakech, Morocco. Antibiotics 2021, 10, 492. [Google Scholar] [CrossRef]
- Jean, S.S.; Lee, W.S.; Hsueh, P.R. Ertapenem non-susceptibility and independent predictors of the carbapenemase production among the Enterobacteriaceae isolates causing intra-abdominal infections in the asia-pacific region: Results from the study for monitoring antimicrobial resistance trends (SMART). Infect. Drug Resist. 2018, 11, 1881–1891. [Google Scholar] [PubMed]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net) Annual Epidemiological Report for 2019. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2019 (accessed on 18 November 2020).
- Ben Helal, R.; Dziri, R.; Chedly, M.; Klibi, N.; Barguellil, F.; El Asli, M.S.; Ben Moussa, M. Occurrence and characterization of carbapenemase-producing Enterobacteriaceae in a Tunisian Hospital. Microb. Drug Resist. 2018, 24, 1361–1367. [Google Scholar] [CrossRef]
- Kim, Y.K.; Chang, I.B.; Kim, H.S.; Song, W.; Lee, S.S. Prolonged Carriage of Carbapenemase-Producing Enterobacteriaceae: Clinical Risk Factors and the Influence of Carbapenemase and Organism Types. J. Clin. Med. 2021, 10, 310. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, M.; Lewtak, K.; Nitsch-Osuch, A. Effectiveness of Antiepidemic Measures Aimed to Reduce Carbapenemase-Producing Enterobacteriaceae in the Hospital Environment. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 9299258. [Google Scholar] [CrossRef]
- Falagas, M.E.; Lourida, P.; Poulikakos, P.; Rafailidis, P.I.; Tansarli, G.S. Antibiotic treatment of infections due to carbapenem-resistant Enterobacteriaceae: Systematic evaluation of the available evidence. Antimicrob. Agents Chemother. 2014, 58, 654–663. [Google Scholar] [CrossRef]
- Chmielewska, S.J.; Leszczyńska, K. Carbapenemase of Intestinal Rods—The Beginning of Post-Antibiotic Era? Adv. Microbiol. 2019, 58, 271–289. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell. Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Tacconelli, E.; Magrini, N. Global Priority List of Antibiotic—Resistant Bacteria to Guide Research, Discovery and Development of New Antibiotics. 2017. Available online: https://www.quotidianosanita.it/allegati/allegato4135670.pdf (accessed on 1 June 2017).
- Mączyńska, B.; Paleczny, J.; Oleksy-Wawrzyniak, M.; Choroszy-Król, I.; Bartoszewicz, M. In Vitro Susceptibility of Multi-Drug Resistant Klebsiella pneumoniae Strains Causing Nosocomial Infections to Fosfomycin. A Comparison of Determination Methods. Pathogens 2021, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Ura, L.; Deja-Makara, B.; Pajdziński, M.; Gottwald, L.M. The occurence and pathogenicity of B-class carbapenemase—Producing Enterobacteriaceae—Klebsiella pneumoniae strains (MBL/NDM) in patients hospitalized and treated in Mazowiecki Memorial Hospital of Radom between 2016–2018. Long-Term Care Nurs. 2020, 3, 239–249. [Google Scholar]
- Qadi, M.; Alhato, S.; Khayyat, R.; Elmanama, A. Colistin Resistance among Enterobacteriaceae Isolated from Clinical Samples in Gaza Strip. Can. J. Infect. Dis. Med. Microbiol. 2021, 2021, 6634684. [Google Scholar] [CrossRef]
- Paul, M.; Daikos, G.L.; Durante-Mangoni, E.; Yahav, D.; Carmeli, Y.; Benattar, Y.D. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: An open-label, randomised controlled trial. Lancet Infect. Dis. 2018, 18, 391–400. [Google Scholar] [CrossRef]
- Ojdana, D.; Gutowska, A.; Sacha, P.; Majewski, P.; Wieczorek, P.; Tryniszewska, E. Activity of Ceftazidime-Avibactam Alone and in Combination with Ertapenem, Fosfomycin, and Tigecycline Against Carbapenemase-Producing Klebsiella pneumoniae. Microb. Drug Resist. 2019, 25, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Medeiros, G.S.; Rigatto, M.H.; Falci, D.R.; Zavascki, A.P. Combination therapy with polymyxin B for carbapenemase-producing Klebsiella pneumoniae bloodstream infection. Int. J. Antimicrob. Agents 2019, 53, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tong, X.; Huang, J.; Zhang, L.; Wang, D.; Wu, M.; Liu, T.; Fan, H. Triple Versus Double Therapy for the Treatment of Severe Infections Caused by Carbapenem-Resistant Enterobacteriaceae: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2020, 10, 1673. [Google Scholar] [CrossRef]
- Galimand, M.; Courvalin, P.; Lambert, T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 2003, 47, 2565–2571. [Google Scholar] [CrossRef]
- Jacoby, G. Mechanism of Resistant to Quinolones. Clin. Inf. Dis. 2005, 41, 5120–5126. [Google Scholar] [CrossRef] [PubMed]
- García-Sureda, L.; Doménech-Sánchez, A.; Barbier, M.; Juan, C.; Gascó, J.; Albertí, S. OmpK26, A novel porin associated with carbapenem resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2011, 55, 4742–4747. [Google Scholar] [CrossRef]
- Padilla, E.; Llobert, E.; Domenech–Sanchez, A.; Alberti, S. Klebsiella pneumoniae AcrAB efflux pump contribute to antimicrobial resustance and virulence. Antimicrob. Agents Chemother. 2009, 10, 1128. [Google Scholar]
- Livermore, D.M.; Mushtaq, S.; Warner, M.; Zhang, J.C.; Maharjan, S.; Doumith, M.; Woodford, N. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J. Antimicrob. Chemother. 2011, 66, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.C.; Burgess, D.S. Treatment of Klebsiella pneumoniae carbapenemase (KPC) infections: A review of published case series and case reports. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, P.; Rodrigues, C.; Shetty, A.; Kapadia, F.; Hedge, A.; Soman, R. New Delhi Metallo-beta lactamase (NDM-1) in Enterobacteriaceae: Treatment options with carbapenems compromised. J. Assoc. Physicians India 2010, 58, 147–149. [Google Scholar] [PubMed]
- Fernando, S.A.; Gray, T.J.; Gottlieb, T. Healthcare-acquired infections: Prevention strategies. Intern. Med. J. 2017, 47, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters; Version 10.0; The European Committee on Antimicrobial Susceptibility Testing: Basel, Switzerland, 2020. [Google Scholar]
- EUCAST. The European Committee on Antimicrobial Susceptibility Testing. Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion as Recommended by EUCAST; Version 10.0; The European Committee on Antimicrobial Susceptibility Testing: Basel, Switzerland, 2020. [Google Scholar]
- Żabicka, D.; Hryniewicz, W. Interpretation Tables of Susceptibility Testing Results According to EUCAST 2018 Recommendations; National Antibiotic Protection Program: Warsaw, Poland, 2020; pp. 24–26.
- Skov, R.; Skov, G. EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. 2017, Volume 6, pp. 1–47. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Resistance_mechanisms/EUCAST_detection_of_resistance_mechanisms_170711.pdf (accessed on 1 June 2022).
- Van Dijk, K.; Voets, G.M.; Scharringa, J.; Voskuil, S.; Fluit, A.C.; Rottier, W.C.; Leverstein-Van Hall, M.A.; Cohen Stuart, J.W. A disc diffusion assay for detection of class A, B and OXA-48 carbapenemases in Enterobacteriaceae using phenyl bo ronic acid, dipicolinic acid and temocillin. Clin. Microbiol. Infect. 2014, 20, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Dortet, L.; Poirel, L.; Errera, C.; Nordmann, P. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J. Clin. Microbiol. 2014, 52, 2359–2364. [Google Scholar] [CrossRef] [PubMed]











| MIC Value (mg/L) | The Size of the Zone Growth Inhibition, Disc 10 µg (mm) | |||
|---|---|---|---|---|
| Antibiotic | S≤ | R> | S≤ | R> |
| Meropenem | 2 | 8 | 22 | 16 |
| Ertapenem | 0.5 | 0.5 | 25 | 25 |
| Imipenem | 2 | 4 | 50 | 17 |
| Name of Starter | Gen | Starter Sequence |
|---|---|---|
| OXA-48a | blaOXA−48 | 5-TTGGTGGCATCGATTATCGG-3 |
| OXA-48b | blaOXA−48 | 5-GAGCACTTCTTTTGTGATGGC-3 |
| KPC-A | blaKP C−2 | 5-CTGTCTTGTCTCTCATGGCC-3 |
| KPC-B | blaKP C−2 | 5-CCTCGCTGTGCTTGTCATCC-3 |
| khe-F | khe | 5-ACCATGTCCGATTTAATCACAACACGC-3 |
| khe-R | khe | 5-GCAGACGAACTTCCTGCTCGGT-3 |
| VIM-UF | blaV IM, blaIMP | 5-GTTTGGTCGCATATCGCAAC-3 |
| VIM-UR | blaV IM | 5-TCAATCTCCGCGAGAAG-3 |
| NDM-F1 | blaNDM | 5-CAGCACACTTCCTATCTCGAC-3 |
| NDM-R1 | blaNDM | 5-GTAGTGCTCAGTGTCGGCATC-3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarowska, J.; Choroszy-Krol, I.; Jama-Kmiecik, A.; Mączyńska, B.; Cholewa, S.; Frej-Madrzak, M. Occurrence and Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Strains Isolated from Hospitalized Patients in Poland—A Single Centre Study. Pathogens 2022, 11, 859. https://doi.org/10.3390/pathogens11080859
Sarowska J, Choroszy-Krol I, Jama-Kmiecik A, Mączyńska B, Cholewa S, Frej-Madrzak M. Occurrence and Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Strains Isolated from Hospitalized Patients in Poland—A Single Centre Study. Pathogens. 2022; 11(8):859. https://doi.org/10.3390/pathogens11080859
Chicago/Turabian StyleSarowska, Jolanta, Irena Choroszy-Krol, Agnieszka Jama-Kmiecik, Beata Mączyńska, Sylwia Cholewa, and Magdalena Frej-Madrzak. 2022. "Occurrence and Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Strains Isolated from Hospitalized Patients in Poland—A Single Centre Study" Pathogens 11, no. 8: 859. https://doi.org/10.3390/pathogens11080859






