O55 Polysaccharides Are Good Antigen Targets for the Formulation of Vaccines against O55 STEC and Capsulated aEPEC Strains
Abstract
1. Introduction
2. Results
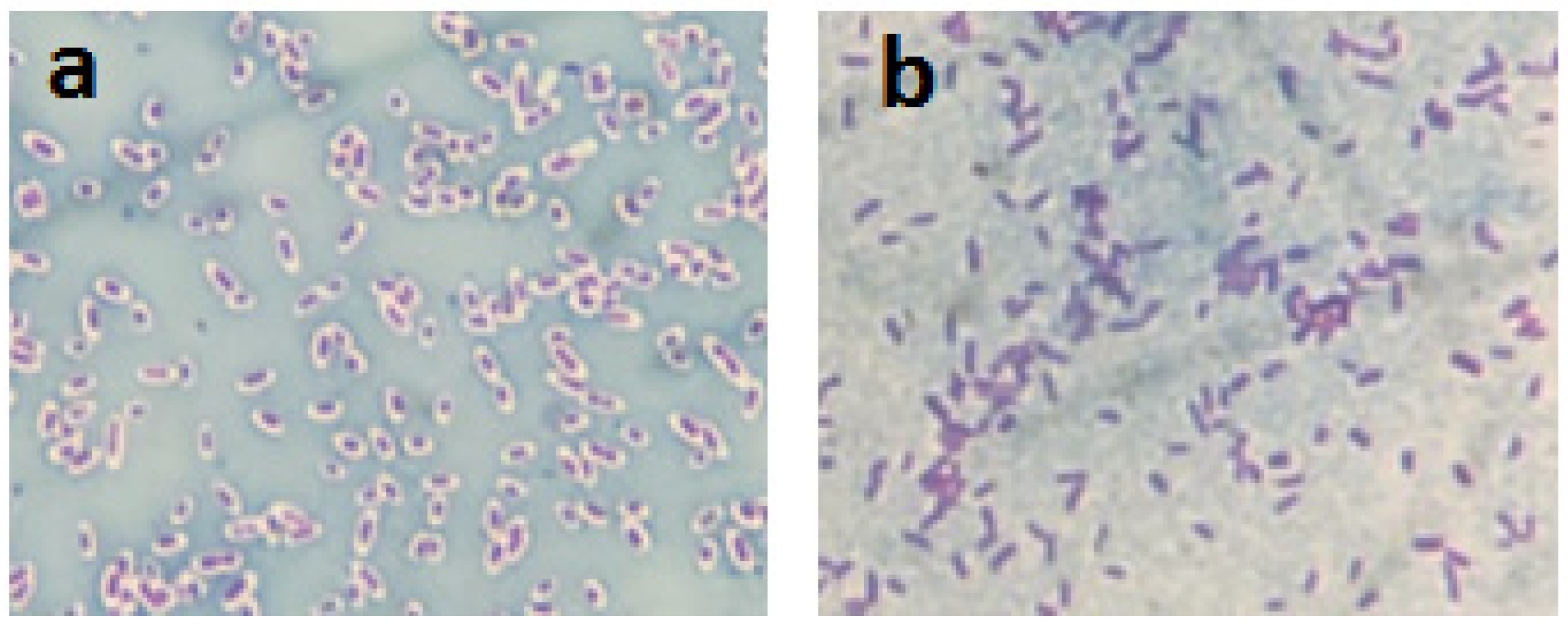
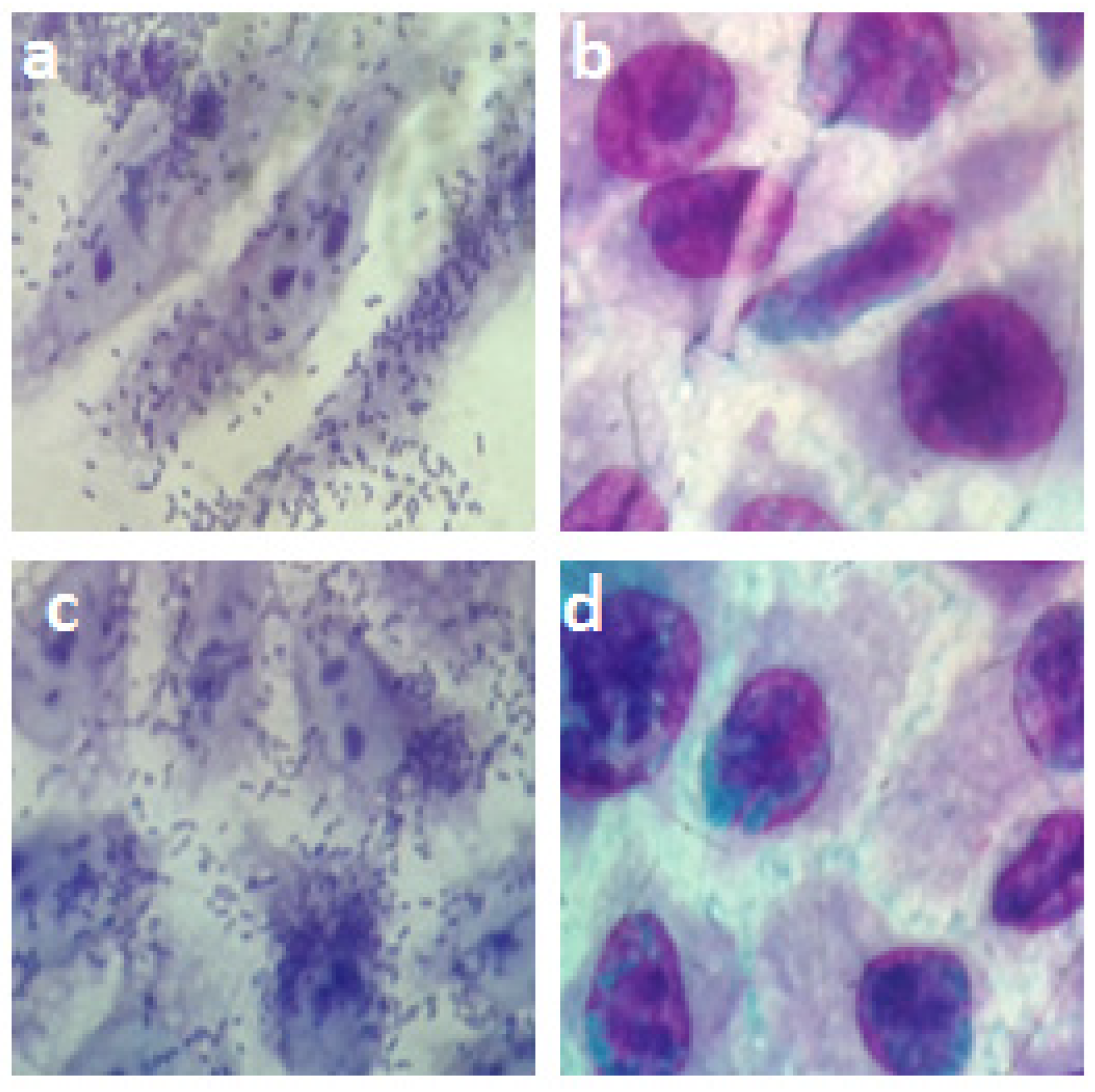
2.1. Visualization of the Bacterial Polysaccharide Capsule
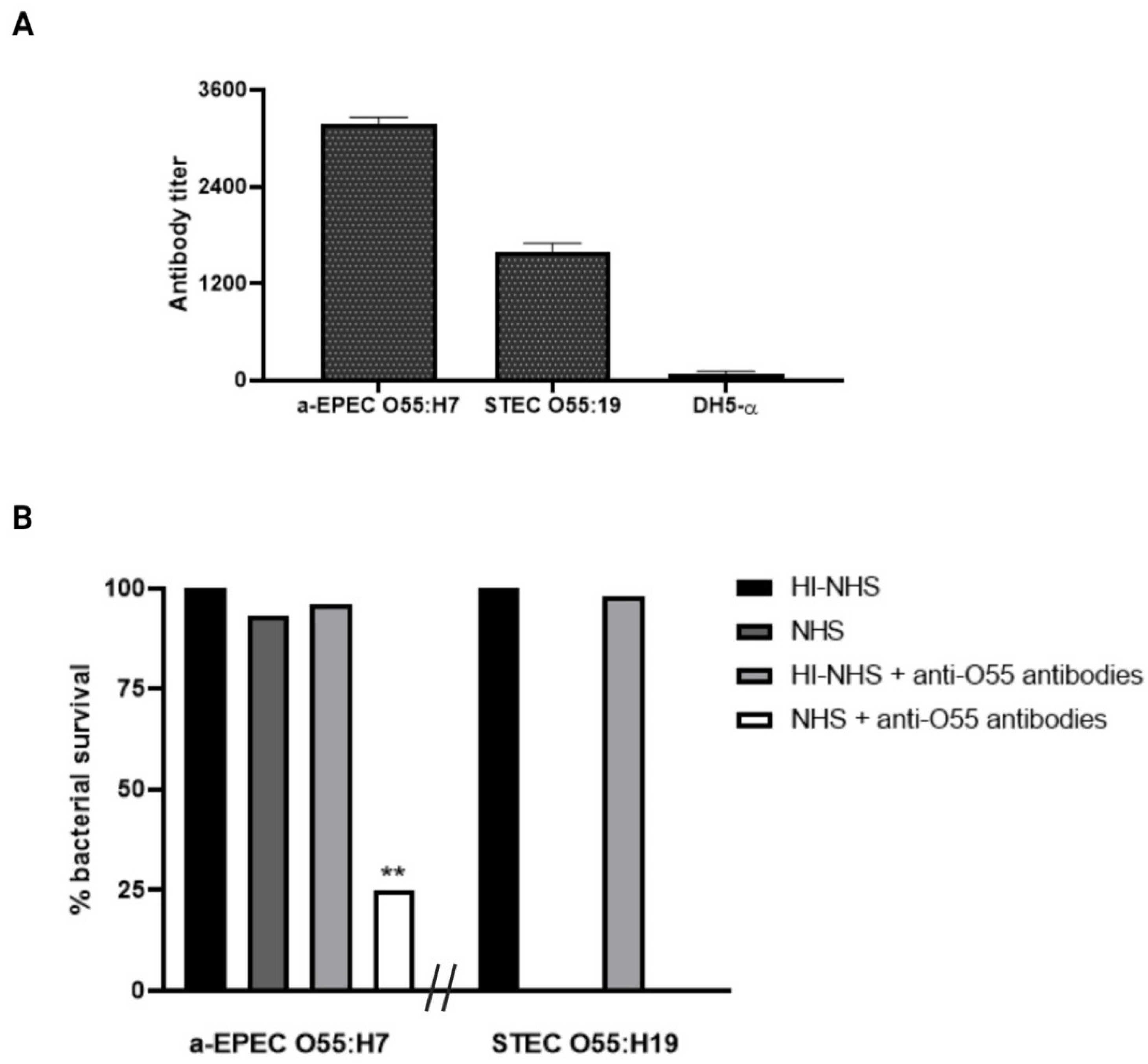
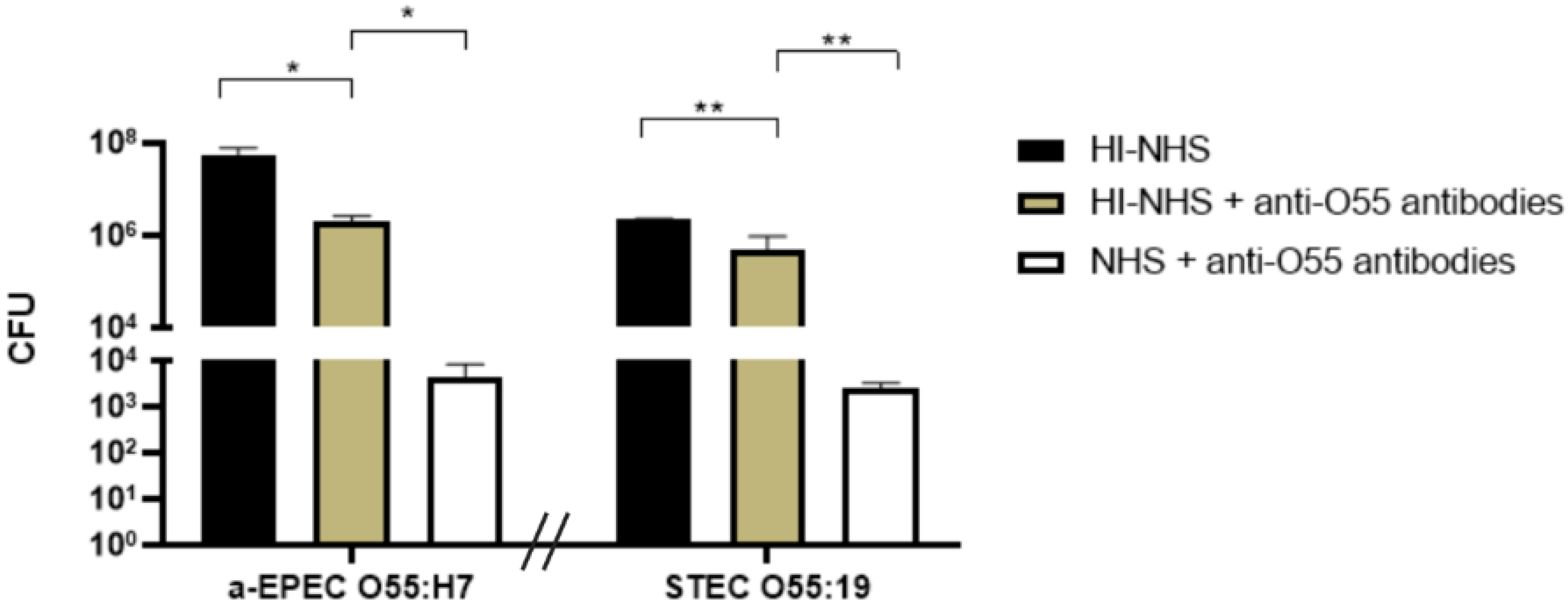
2.2. Influence of Anti-O55 Antibodies in Bacterial Recognition and Complement Response
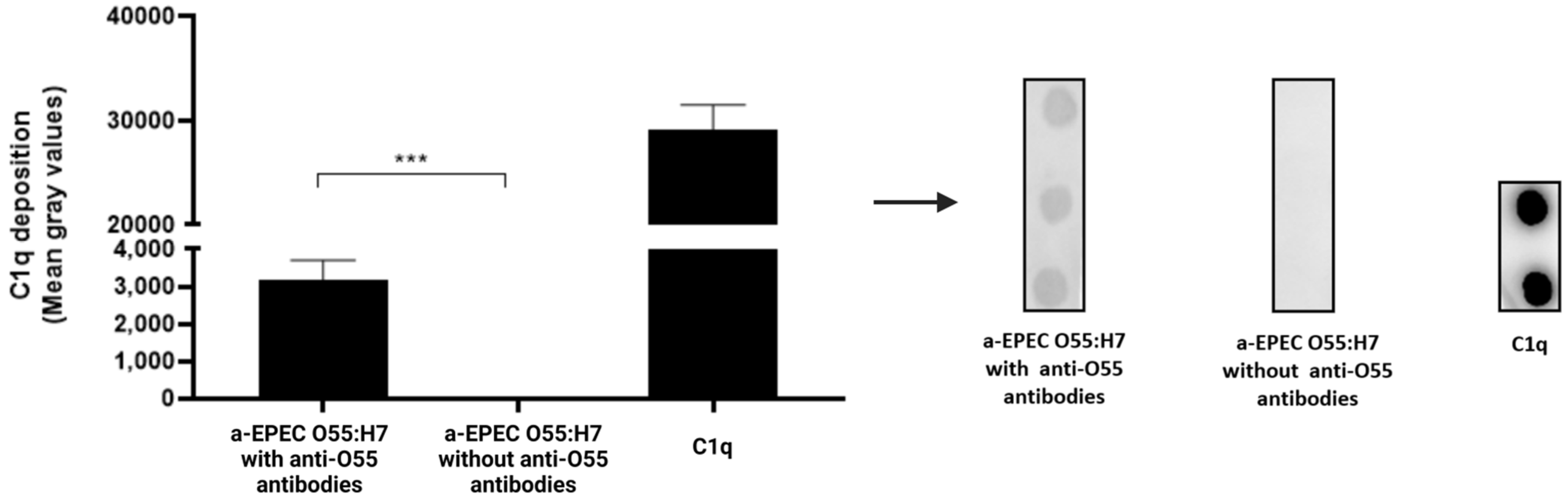
2.3. Deposition of C3b and C1q on the Bacterial Surface
2.4. Influence of Anti-O55 Antibodies in Phagocytosis and Bacterial Adhesion
3. Discussion
4. Material and Methods
4.1. Bacterial Strains
4.2. Cell Lines
4.3. Anti-O55 Antibodies
4.4. Capsule Visualization
4.5. Anti-O55 Antibody Detection
4.6. Complement Lysis
4.7. C3b Deposition on the Bacterial Surface
4.8. C1q Deposition on the Bacterial Surface
4.9. Phagocytosis
4.10. Inhibition of Bacterial Adhesion to Epithelial Cells
4.11. Statistical Analysis
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). Available online: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease (accessed on 1 August 2022).
- Merino, V.R.; Nakano, V.; Delannoy, S.; Fach, P.; Alberca, G.G.F.; Farfan, M.J.; Piazza, R.M.F.; Avila-Campos, M.J. Prevalence of Enteropathogens and virulence traits in Brazilian children with or without diarrhea. Front. Cell. Infect. Microbiol. 2020, 10, 549919. [Google Scholar] [CrossRef] [PubMed]
- Gaytan, M.O.; Martinez-Santos, V.I.; Soto, E.; Gonzales-Pedrajo, B. Type three secretion system in attaching and effecing pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 129. [Google Scholar] [CrossRef]
- Campos, L.C.; Franzolin, M.R.; Trabulsi, L.R. Diarrheagenic Escherichia coli Categories among the Traditional Enteropathogenic E. coli O Serogroups—A Review. Mem. Inst. Oswaldo Cruz. 2004, 99, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Perepelov, A.V.; Guo, X.; Cao, H.; Wang, Q.; Reeves, P.R.; Knirel, Y.A.; Wang, L.; Widmalm, G. Structure and genetics of Escherichia coli O antigens. FEMS Microbiol. Rev. 2020, fuz28, 655–683. [Google Scholar] [CrossRef] [PubMed]
- Bosilevec, J.M.; Koohmaraie, M. Prevalence and characterization of non-O157 shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl. Environ. Microbiol. 2011, 77, 2103–2112. [Google Scholar] [CrossRef]
- Gomes, T.A.T.; Irino, K.; Girão, D.M.; Girão, V.B.C.; Guth, B.E.C.; Vaz, T.M.I.; Moreira, F.C.; Chinarelli, S.H.; Vieira, M.A.M. Emerging enteropathogenic Escherichia coli strains. Emerg. Infec. Dis. 2004, 10, 1851–1855. [Google Scholar] [CrossRef]
- Vaz, T.M.I.; Irino, K.; Kato, M.A.M.; Dias, A.M.G.; Gomes, T.A.T.; Medeiros, M.I.C.; Rocha, M.M.M.; Guth, B.E.C. Virulence properties and characteristic of shiga toxin-producing Escherichia coli in São Paulo, Brazil, from 1976 through 1999. J. Clin. Microbiol. 2004, 2, 903–905. [Google Scholar] [CrossRef] [PubMed]
- Gomes, T.A.T.; Elias, W.P.; Scaletsky, I.C.A.; Guth, B.E.C.; Rodrigues, J.F.; Piazza, R.M.F.; Ferreira, L.C.S.; Martinez, M.B. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016, 47, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, C.; Vishram, B.; Jenkins, C.; Jorgensen, F.; Byrne, L.; Mikhail, A.F.W.; Dallman, T.J.; Carroll, K.; Ahyow, L.; Vahora, Q.; et al. Epidemiological investigation of recurrent outbreaks of hemolytic uremic syndrome caused by Shiga toxin-producing Escherichia coli. Epidemiol. Infect. 2021, 149, e108. [Google Scholar] [CrossRef] [PubMed]
- Wick, L.M.; Qi, W.; Lacher, D.W.; Whittam, T.S. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 2005, 187, 1783–1791. [Google Scholar] [CrossRef] [PubMed]
- McFarland, N.; Bundle, N.; Jenkins, C.; Godbole, G.; Mikhail, A.; Dallman, T.; O’Connor, C.; McCarthy, N.; O’Connell, E.; Treacy, J.; et al. Recurrent seasonal outbreak of an emerging serotype of Shiga toxin-producing Escherichia coli (STEC O55:H7 Stx2a) in the south west of England. Euro Surveill. 2017, 36, 30610. [Google Scholar] [CrossRef]
- Whitfield, C. Biosynthesis and Assembly of Capsular Polysaccharides in Escherichia coli. Annu. Rev. Biochem. 2006, 75, 39–68. [Google Scholar] [CrossRef] [PubMed]
- Chowers, Y.; Kirschner, J.; Keller, N.; Barshack, I.; Bar-Meir, S.; Ashkenari, S.; Schneerson, R.; Robbins, J.; Passwell, J. O-specific polysaccharide conjugate vaccine-induced IgG antibodies prevent invasion of Shigella into Caco-2 cells and may be curative. Proc. Natl. Acad. Sci. USA 2007, 104, 2396–2401. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.F.; New, R.R.C.; Andrade, G.R.; Ozaki, C.Y.; Sant’Anna, O.A.; Mendonça-Previato, L.; Trabulsi, L.R.; Domingos, M.O. Lipopolysaccharide as an antigen Target for the Formulation of a Universal Vaccine against Escherichia coli O111 strains. Clin. Vaccine Immul. 2010, 11, 1172–1780. [Google Scholar] [CrossRef]
- Andrade, G.R.; New, R.R.C.; Sant’Anna, O.A.; Williams, N.A.; Alves, R.C.B.; Pimenta, D.C.; Viferelli, H.; Melo, B.S.; Rocha, L.B.; Piazza, R.M.F.; et al. A universal polysaccharide conjugated vaccine against O111 E. coli. Hum. Vaccines Immunother. 2014, 10, 2864–2874. [Google Scholar] [CrossRef]
- Ledov, V.; Golovina, M.E.; Markina, A.A.; Knirel, Y.A.; Lvov, V.L.; Kovalchuck, A.L.; Aparin, P.G. Highly homogenous tri-acylated S-LPS act as a novel clinically applicable vaccine against Shigella flexneri 2a infection. Vaccine 2019, 37, 1062–1972. [Google Scholar] [CrossRef]
- Perera, S.R.; Sokaribo, A.S.; White, A.P. Polysaccharide Vaccines: A perspective on non-typhoidal Salmonella. Polysaccharides 2021, 2, 691–714. [Google Scholar] [CrossRef]
- Elsheimer-Matulova, M.; Aribam, S.D.; Nkayama, M.; Ogawa, Y.; Shimoji, Y.; Eguchi, M. The protective capacity of anti-O4 antigen antibodies against Salmonella infection is influenced by the presence or absence of the O5 antigen. Vaccine 2020, 38, 5408–5412. [Google Scholar] [CrossRef]
- Whitfield, C. Structure and Assembly of Escherichia coli Capsules. In Cellular and Molecular Biology of E. coli, Salmonella, and the Enterobacteriacea; Sauch, J.M., Ed.; The School of Molecular Biology, University of Illinois at Urbana-Champaign: Champaign, IL, USA, 2009. [Google Scholar] [CrossRef]
- Wen, Z.; Zang, J.R. Bacterial capsules. In Molecular Medical Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2015; Chapter 3; Volume 1, pp. 33–35. [Google Scholar] [CrossRef]
- Reis, E.S.; Mastellos, D.C.; Hajishengallis, G.; Lambris, J.D. New insights into the immune functions of complement. Nat. Rev. Immunol. 2019, 19, 503–516. [Google Scholar] [CrossRef]
- Biran, D.; Rosenshine, I.; Ron, E.Z. Escherichia coli O-antigen capsule (group 4) is essential for serum resistance. Res. Microbiol. 2020, 171, 99–101. [Google Scholar] [CrossRef]
- Abreu, A.G.; Barbosa, A.S. How Escherichia coli circumvet complement-mediated killing. Front. Immunol. 2017, 8, 452. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ord, A.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; Lindqvist, R.; et al. Pathogenicity assessment of Shiga toxin-producing Escherichia coli (STEC) and the public health risk posed by contamination of food with STEC. EFSA J. 2020, 18, 5967. [Google Scholar] [CrossRef]
- Torti, J.F.; Cuervo, P.; Nardelo, A.; Pizarro, M. Epidemiology and Characterization of Shiga Toxin-Producing Escherichia coli of Hemolytic Uremic Syndrome in Argentina. Cureus 2021, 13, e17213. [Google Scholar] [CrossRef]
- Toshima, H.; Yoshimura, A.; Arikawa, K.; Hidaka, A.; Ogasawara, J.; Hase, A.; Masaki, H.; Nishikawa, Y. Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl. Environ. Microbiol. 2007, 73, 7582–7588. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kimmitt, P.T.; Harwood, C.R.; Barer, M.R. Toxin gene expression by Shiga toxin–producing Escherichia coli: The role of antibiotics and the bacterial SOS response. Emerg. Infect Dis. 2000, 6, 458–465. [Google Scholar] [CrossRef]
- Davis, T.K.; McKee, R.; Schnadower, D.; Tarr, P.I. Treatment of Shiga toxin–producing Escherichia coli infections. Infect Dis. Clin. N. Am. 2013, 27, 577–597. [Google Scholar] [CrossRef] [PubMed]
- Fingermann, M.; Avila, L.; De Marco, M.B.; Vazquez, L.; Di Biase, D.M.; Muller, A.V.; Lescano, M.; Dokmetjian, C.; Castillo, S.F.; Quinoy, J.L.P. OMV-based vaccine formulations against SHiga toxin producing Escherichia coli strains are both protective in mice and immunogenic in calves. Hum. Vaccines Immunoterapeutics 2018, 14, 2208–2213. [Google Scholar] [CrossRef] [PubMed]
- Larzábal, M.; Cataldi, A.A.; Vilte, D.A. Human and Veterinary vaccines against pathogenic Escherichia coli. In The Universe of Escherichia coli.; Erjavec, M.S., Ed.; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Henrique, I.M.; Sacerdoti, F.; Ferreira, R.L.; Henrique, C.; Amaral, M.M.; Piazza, R.M.F.; Luz, D. Therapeutic antibodies against Shiga Toxins: Trends and Perspectives. Front. Cell Infect. Microbiol. 2022, 12, 825856. [Google Scholar] [CrossRef] [PubMed]
- Nesta, B.; Pizza, M. Vaccines against Escherichia coli. Curr. Top. Microbiol. Immunol. 2018, 416, 213–242. [Google Scholar] [CrossRef]
- Rodrigues, J.; Scaletsky, I.C.A.; Campos, L.C.; Gomes, T.A.T.; Whittam, T.S.; Trabulsi, L.R. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect. Immun. 1996, 64, 2680–2686. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.A.; Santos, A.R.R.; Rocha, L.B.; Caetano, B.A.; Mitsunari, T.; Santos, L.I.; Polatto, J.M.; Horton, D.S.P.Q.; Guth, B.E.C.; Santos, L.F.; et al. Development and Validation of shiga toxin-producing Escherichia coli. Immunodiagnostic assay. Microorganisms 2019, 7, 276. [Google Scholar] [CrossRef] [PubMed]
- Hughes, R.; Smith, A.C. Capsule Stain Protocols. American Society for Microbiology. 2007. Available online: https://asm.org/Protocols/Capsule-Stain-Protocols (accessed on 4 April 2022).
- Baron, F.; Cochet, M.F.; Ablain, W.; Grosset, N.; Madec, M.N.; Gonnet, F.; Jan, S.; Gautier, M. Rapid and cost-effective method for micro-organism enumeration based on miniaturization of conventional plate-couting technique. Lait INRA Ed. 2006, 86, 251–257. [Google Scholar] [CrossRef]
- Beck, N.K.; Callahan, K.; Nappier, S.P.; Kim, H.; Sobsey, M.D.; Meschke, J.S. Development of a Spot-Titer Culture assay for Quantifying bacteria and viral. J. Rapid Methods Autom. Microbiol. 2009, 17, 455–464. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, H.G.d.S.; Franzolin, M.R.; Anjos, G.F.d.; Barbosa, A.S.; Santos, L.F.d.; Miranda, K.F.; Marques, R.M.; Souza, M.C.L.d.; Piazza, R.M.F.; Domingos, M.d.O. O55 Polysaccharides Are Good Antigen Targets for the Formulation of Vaccines against O55 STEC and Capsulated aEPEC Strains. Pathogens 2022, 11, 895. https://doi.org/10.3390/pathogens11080895
Silva HGdS, Franzolin MR, Anjos GFd, Barbosa AS, Santos LFd, Miranda KF, Marques RM, Souza MCLd, Piazza RMF, Domingos MdO. O55 Polysaccharides Are Good Antigen Targets for the Formulation of Vaccines against O55 STEC and Capsulated aEPEC Strains. Pathogens. 2022; 11(8):895. https://doi.org/10.3390/pathogens11080895
Chicago/Turabian StyleSilva, Herbert Guimarães de Sousa, Marcia Regina Franzolin, Geovana Ferreira dos Anjos, Angela Silva Barbosa, Luis Fernando dos Santos, Kaique Ferrari Miranda, Ronaldo Maciel Marques, Matilde Costa Lima de Souza, Roxane Maria Fontes Piazza, and Marta de Oliveira Domingos. 2022. "O55 Polysaccharides Are Good Antigen Targets for the Formulation of Vaccines against O55 STEC and Capsulated aEPEC Strains" Pathogens 11, no. 8: 895. https://doi.org/10.3390/pathogens11080895
APA StyleSilva, H. G. d. S., Franzolin, M. R., Anjos, G. F. d., Barbosa, A. S., Santos, L. F. d., Miranda, K. F., Marques, R. M., Souza, M. C. L. d., Piazza, R. M. F., & Domingos, M. d. O. (2022). O55 Polysaccharides Are Good Antigen Targets for the Formulation of Vaccines against O55 STEC and Capsulated aEPEC Strains. Pathogens, 11(8), 895. https://doi.org/10.3390/pathogens11080895






