Faecal Carriage of Carbapenem-Resistant Acinetobacter baumannii: Comparison to Clinical Isolates from the Same Period (2017–2019)
Abstract
:1. Introduction
2. Results
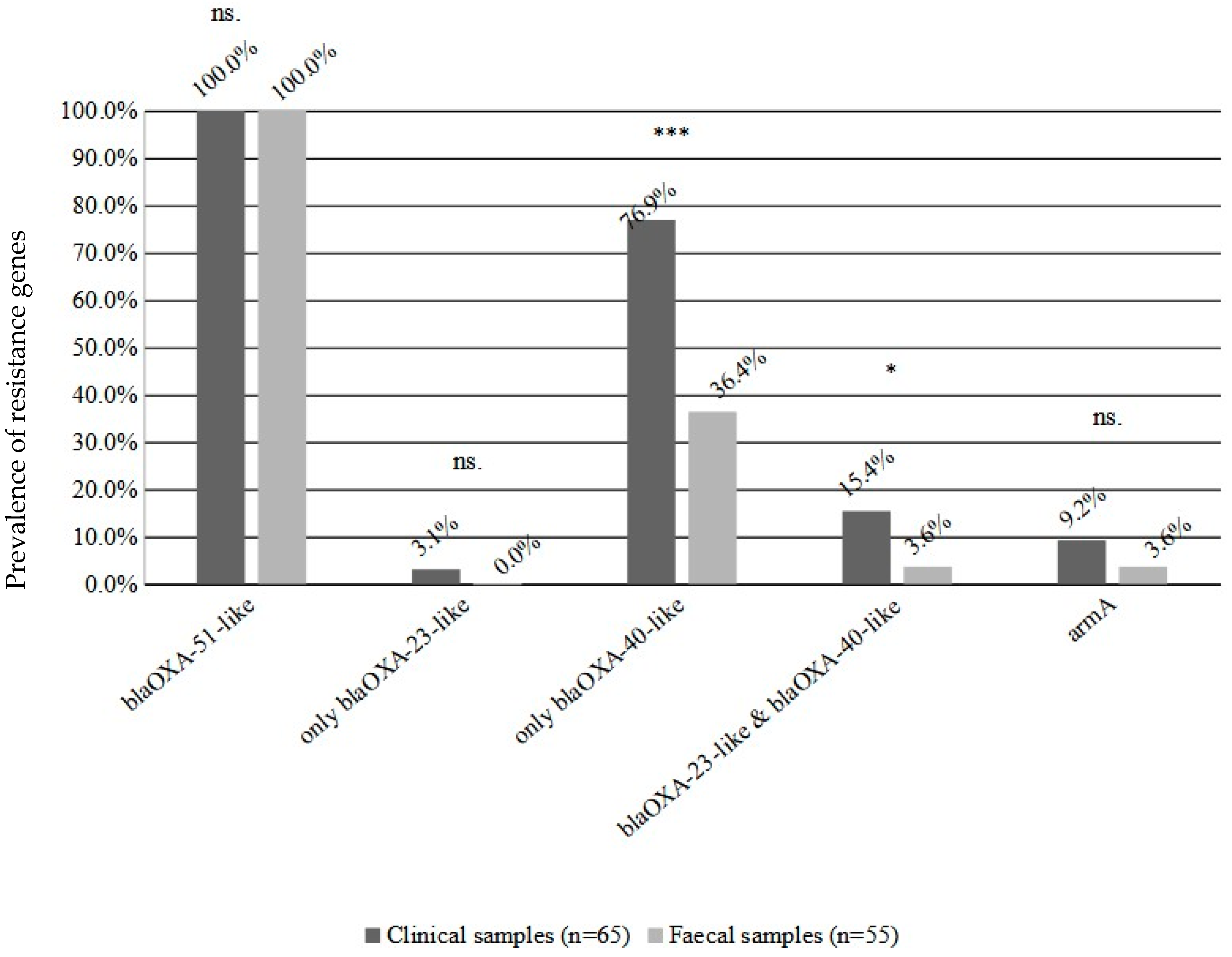
2.1. Prevalence, Susceptibility Testing and Resistance Genes
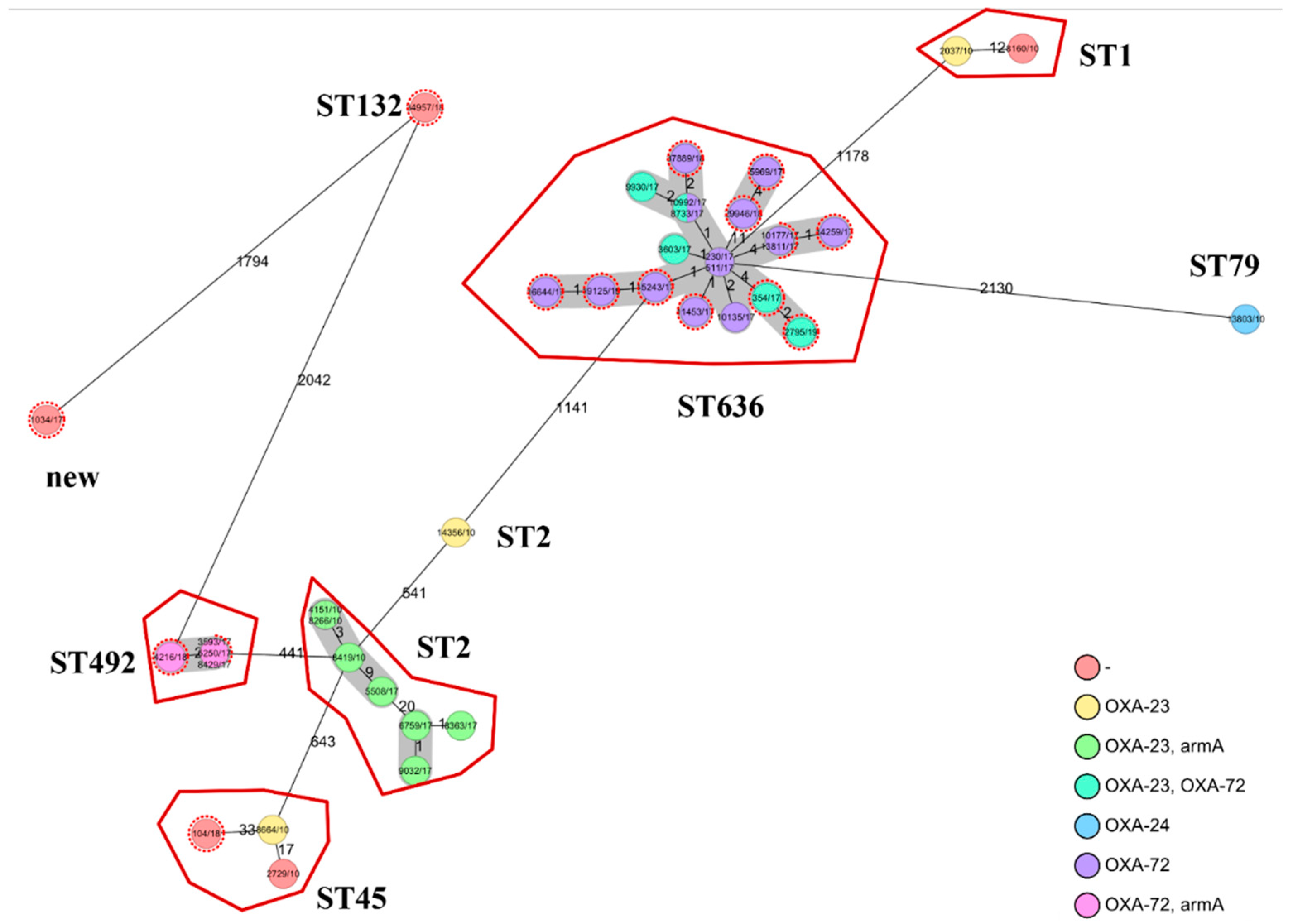
2.2. Whole Genome Sequencing (WGS)
2.3. Comparison of Faecal and Clinical Isolates
3. Discussion
4. Materials and Methods
4.1. Isolates
4.2. Resistance Genes
4.3. Whole Genome Sequencing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and caveats in combating ESKAPE pathogens against nosocomial infections. Adv. Sci. 2020, 7, 1901872. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling a Crisis for the Health and Wealth of Nations. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crsis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf (accessed on 24 March 2019).
- Shrestha, P.; Cooper, B.S.; Coast, J.; Oppong, R.; Do Thi Thuy, N.; Phodha, T.; Celhay, O.; Guerin, P.J.; Wertheim, H.; Lubell, Y. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control. 2018, 7, 98. [Google Scholar] [CrossRef]
- Mehta, Y.; Jaggi, N.; Rosenthal, V.D.; Kavathekar, M.; Sakle, A.; Munshi, N.; Chakravarthy, M.; Todi, S.K.; Saini, N.; Rodrigues, C.; et al. Device-Associated Infection Rates in 20 Cities of India, Data Summary for 2004–2013: Findings of the International Nosocomial Infection Control Consortium. Infect. Control. Hosp. Epidemiol. 2016, 37, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Y.; Lyles-Banks, R.D.; Lolans, K.; Hines, D.W.; Spear, J.B.; Petrak, R.; Trick, W.E.; Weinstein, R.A.; Hayden, M.K.; Centers for Disease Control and Prevention Epicenters Program. The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2013, 57, 1246–1252. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.K.; Lin, M.Y.; Lolans, K.; Weiner, S.; Blom, D.; Moore, N.M.; Fogg, L.; Henry, D.; Lyles, R.; Thurlow, C.; et al. Prevention of colonization and infection by Klebsiella pneumoniae carbapenemase-producing enterobacteriaceae in long-term acute-care hospitals. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2015, 60, 1153–1161. [Google Scholar] [CrossRef]
- Shimasaki, T.; Seekatz, A.; Bassis, C.; Rhee, Y.; Yelin, R.D.; Fogg, L.; Dangana, T.; Cisneros, E.C.; Weinstein, R.A.; Okamoto, K.; et al. Increased Relative Abundance of Klebsiella pneumoniae Carbapenemase-producing Klebsiella pneumoniae Within the Gut Microbiota Is Associated With Risk of Bloodstream Infection in Long-term Acute Care Hospital Patients. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2019, 68, 2053–2059. [Google Scholar] [CrossRef]
- Juhász, J.; Ligeti, B.; Gajdács, M.; Makra, N.; Ostorházi, E.; Farkas, F.B.; Stercz, B.; Tóth, Á.; Domokos, J.; Pongor, S.; et al. Colonization Dynamics of Multidrug-Resistant Klebsiella pneumoniae Are Dictated by Microbiota-Cluster Group Behavior over Individual Antibiotic Susceptibility: A Metataxonomic Analysis. Antibiotics 2021, 10, 268. [Google Scholar] [CrossRef]
- Lob, S.H.; Biedenbach, D.J.; Badal, R.E.; Kazmierczak, K.M.; Sahm, D.F. Antimicrobial resistance and resistance mechanisms of Enterobacteriaceae in ICU and non-ICU wards in Europe and North America: SMART 2011–2013. J. Glob. Antimicrob. Resist. 2015, 3, 190–197. [Google Scholar] [CrossRef]
- Karanika, S.; Karantanos, T.; Arvanitis, M.; Grigoras, C.; Mylonakis, E. Fecal Colonization With Extended-spectrum Beta-lactamase-Producing Enterobacteriaceae and Risk Factors Among Healthy Individuals: A Systematic Review and Metaanalysis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2016, 63, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Wu, S.; Hao, M.; Zhu, J.; Ding, B.; Yang, Y.; Xu, X.; Wang, M.; Yang, F.; Hu, F. The Colonization of Carbapenem-Resistant Klebsiella pneumoniae: Epidemiology, Resistance Mechanisms, and Risk Factors in Patients Admitted to Intensive Care Units in China. J. Infect. Dis. 2020, 221, S206–S214. [Google Scholar] [CrossRef] [PubMed]
- Aljindan, R.; Bukharie, H.; Alomar, A.; Abdalhamid, B. Prevalence of digestive tract colonization of carbapenem-resistant Acinetobacter baumannii in hospitals in Saudi Arabia. J. Med. Microbiol. 2015, 64 Pt 4, 400–406. [Google Scholar] [CrossRef]
- Mózes, J.; Ebrahimi, F.; Gorácz, O.; Miszti, C.; Kardos, G. Effect of carbapenem consumption patterns on the molecular epidemiology and carbapenem resistance of Acinetobacter baumannii. J. Med. Microbiol. 2014, 63, 1654–1662. [Google Scholar] [CrossRef]
- Balázs, B.; Tóth, Z.; Nagy, F.; Kovács, R.; Tóth, H.; Nagy, J.B.; Tóth, Á.; Szarka, K.; Majoros, L.; Kardos, G. The Role of Uniform Meropenem Usage in Acinetobacter baumannii Clone Replacement. Antibiotics 2021, 10, 127. [Google Scholar] [CrossRef]
- Corbella, X.; Pujol, M.; Ayats, J.; Sendra, M.; Ardanuy, C.; Dominguez, M.A.; Linares, J.; Ariza, J.; Gudiol, F. Relevance of digestive tract colonization in the epidemiology of nosocomial infections due to multiresistant Acinetobacter baumannii. Clin. Infect. Dis. 1996, 23, 329–334. [Google Scholar] [CrossRef]
- Maamar, E.; Alonso, C.A.; Ferjani, S.; Jendoubi, A.; Hamzaoui, Z.; Jebri, A.; Saisani, M.; Ghedira, S.; Torres, C.; Boubaker, I.B.B. NDM-1-and OXA-23-producing Acinetobacter baumannii isolated from intensive care unit patients in Tunisia. Int. J. Antimicrob. Agents 2018, 52, 910–915. [Google Scholar] [CrossRef]
- Dijkshoorn, L.; Van Aken, E.; Shunburne, L.; Van Der Reijden, T.J.K.; Bernards, A.T.; Nemec, A.; Towner, K.J. Prevalence of Acinetobacter baumannii and other Acinetobacter spp. in faecal samples from non-hospitalised individuals. Clin. Microbiol. Infect. 2005, 11, 329–332. [Google Scholar] [CrossRef]
- Li, S.; Duan, X.; Peng, Y.; Rui, Y. Molecular characteristics of carbapenem-resistant Acinetobacter spp. from clinical infection samples and fecal survey samples in Southern China. BMC Infect. Dis. 2019, 19, 900. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Mózes, J.; Monostori, J.; Gorácz, O.; Fésűs, A.; Majoros, L.; Szarka, K.; Kardos, G. Comparison of rates of fecal colonization with extended-spectrum beta-lactamase-producing enterobacteria among patients in different wards, outpatients and medical students. Microbiol. Immunol. 2016, 60, 285–294. [Google Scholar] [CrossRef] [Green Version]
- Wohrley, J.D.; Bartlett, A.H. The Role of the Environment and Colonization in Healthcare-Associated Infections. Healthc.-Assoc. Infect. Child. 2018, 16, 17–36. [Google Scholar]
- Petrović, T.; Uzunović, S.; Barišić, I.; Luxner, J.; Grisold, A.; Zarfel, G.; Ibrahimagić, A.; Jakovac, S.; Slaćanac, D.; Bedenić, B. Arrival of carbapenem-hydrolyzing-oxacillinases in Acinetobacter baumannii in Bosnia and Herzegovina. Infect. Genet. Evol. 2018, 58, 192–198. [Google Scholar] [PubMed]
- Nemec, A.; Krizova, L.; Maixnerova, M.; Musilek, M. Multidrug-resistant epidemic clones among bloodstream isolates of Pseudomonas aeruginosa in the Czech Republic. Res. Microbiol. 2010, 161, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Villalón, P.; Ortega, M.; Sáez-Nieto, J.A.; Carrasco, G.; Medina-Pascual, M.J.; Garrido, N.; Valdezate, S. Dynamics of a sporadic nosocomial Acinetobacter calcoaceticus–Acinetobacter baumannii complex population. Front. Microbiol. 2019, 10, 593. [Google Scholar] [PubMed]
- Gheorghe, I.; Barbu, I.C.; Surleac, M.; Sârbu, I.; Popa, L.I.; Paraschiv, S.; Feng, Y.; Lazar, V.; Chifiriuc, M.C.; Oţelea, D.; et al. Subtypes, resistance and virulence platforms in extended-drug resistant Acinetobacter baumannii Romanian isolates. Sci. Rep. 2021, 11, 13288. [Google Scholar]
- Gniadek, T.J.; Carroll, K.C.; Simner, P.J. Carbapenem-resistant non-glucose-fermenting Gram-negative bacilli: The missing piece to the puzzle. J. Clin. Microbiol. 2016, 54, 1700–1710. [Google Scholar]
- Weinberg, S.E.; Villedieu, A.; Bagdasarian, N.; Karah, N.; Teare, L.; Elamin, W.F. Control and management of multidrug resistant Acinetobacter baumannii: A review of the evidence and proposal of novel approaches. Infect. Prev. Pract. 2020, 2, 100077. [Google Scholar]
- Pachori, P.; Gothalwal, R.; Gandhi, P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019, 6, 109–119. [Google Scholar]
- Gijón, D.; Curiao, T.; Baquero, F.; Coque, T.M.; Cantón, R. Fecal carriage of carbapenemase-producing Enterobacteriaceae: A hidden reservoir in hospitalized and nonhospitalized patients. J. Clin. Microbiol. 2012, 50, 1558–1563. [Google Scholar] [CrossRef]
- Available online: https://www.eucast.org/ast_of_bacteria/previous_versions_of_documents/ (accessed on 1 January 2018).
- Anil, S. Granger causality. Scholarpedia 2007, 2, 1667. [Google Scholar]
- Wessa, P. Bivariate Granger Causality (v1.0.4) in Free Statistics Software (v1.2.1), Office for Research Development and Education. 2016. Available online: http://www.wessa.net/rwasp_grangercausality.wasp/ (accessed on 1 January 2022).
- Naas, T.; Levy, M.; Hirschauer, C.; Marchandin, H.; Nordmann, P. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-23 in a tertiary care hospital of Papeete, French Polynesia. J. Clin. Microbiol. 2005, 43, 4826–4829. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, P.; Galimand, M.; Bauraing, C.; Deplano, A.; Vanhoof, R.; De Mendonca, R.; Rodriguez-Villalobos, H.; Struelens, M.; Glupczynski, Y. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 2007, 59, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Isolate | Year | ST | Acquired CHDLs | Resistance Phenotype | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | COL | CIP | AMN | GMN | TMN | ||||
| ab61 Ac.No.: SRX14056986 | 2017 | new | None | S | S | S | R | S | S | S |
| ab64 Ac.No.: SRX14056995 | 2017 | 492 | blaOXA-72 | R | R | S | R | R | R | R |
| ab65 Ac.No.: SRX14056996 | 2017 | 636 | blaOXA-72 | R | R | S | R | S | R | S |
| ab66 Ac. No.: SRX14056997 | 2017 | 636 | blaOXA-72 | S | S | S | R | R | S | R |
| ab67 Ac.No.: SRX14056998 | 2017 | 636 | blaOXA-72 | R | R | S | R | S | R | S |
| ab68 Ac.No.: SRX14056999 | 2017 | 636 | blaOXA-72 | R | R | S | R | S | S | S |
| ab69 Ac.No.: SRX14057000 | 2017 | 636 | blaOXA-72 | R | R | S | S | S | S | S |
| ab71 Ac.No.: SRX14056988 | 2017 | 636 | blaOXA-72 | R | R | S | R | S | R | S |
| ab72 Ac.No.: SRX14056989 | 2017 | 636 | blaOXA-23 blaOXA-72 | R | R | S | R | R | R | R |
| ab73 Ac.No.: SRX14056990 | 2017 | 636 | blaOXA-72 | R | R | S | R | S | R | S |
| ab74 Ac.No.: SRX14056991 | 2017 | 636 | blaOXA-72 | R | R | S | R | R | R | R |
| ab75 Ac.No.: SRX14056992 | 2017 | 636 | blaOXA-72 | R | R | S | R | S | R | S |
| ab60 Ac.No.: SRX14056985 | 2018 | 45 | None | S | S | S | R | R | S | R |
| ab63 Ac.No.: SRX14056994 | 2018 | 492 | blaOXA-72 | R | R | S | R | R | R | R |
| ab70 Ac.No.: SRX14056987 | 2018 | 132 | None | S | S | S | S | S | S | S |
| ab62 Ac.No.: SRX14056993 | 2019 | 636 | blaOXA-23 blaOXA-72 | R | R | S | R | R | R | R |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balázs, B.; Tóth, Z.; Nagy, J.B.; Majoros, L.; Tóth, Á.; Kardos, G. Faecal Carriage of Carbapenem-Resistant Acinetobacter baumannii: Comparison to Clinical Isolates from the Same Period (2017–2019). Pathogens 2022, 11, 1003. https://doi.org/10.3390/pathogens11091003
Balázs B, Tóth Z, Nagy JB, Majoros L, Tóth Á, Kardos G. Faecal Carriage of Carbapenem-Resistant Acinetobacter baumannii: Comparison to Clinical Isolates from the Same Period (2017–2019). Pathogens. 2022; 11(9):1003. https://doi.org/10.3390/pathogens11091003
Chicago/Turabian StyleBalázs, Bence, Zoltán Tóth, József Bálint Nagy, László Majoros, Ákos Tóth, and Gábor Kardos. 2022. "Faecal Carriage of Carbapenem-Resistant Acinetobacter baumannii: Comparison to Clinical Isolates from the Same Period (2017–2019)" Pathogens 11, no. 9: 1003. https://doi.org/10.3390/pathogens11091003
APA StyleBalázs, B., Tóth, Z., Nagy, J. B., Majoros, L., Tóth, Á., & Kardos, G. (2022). Faecal Carriage of Carbapenem-Resistant Acinetobacter baumannii: Comparison to Clinical Isolates from the Same Period (2017–2019). Pathogens, 11(9), 1003. https://doi.org/10.3390/pathogens11091003





