Detection and Multigene Typing of ‘Candidatus Phytoplasma solani’-Related Strains Infecting Tomato and Potato Plants in Different Regions of Turkey
Abstract
:1. Introduction
2. Results
2.1. Phytoplasma Detection and Symptom Observation
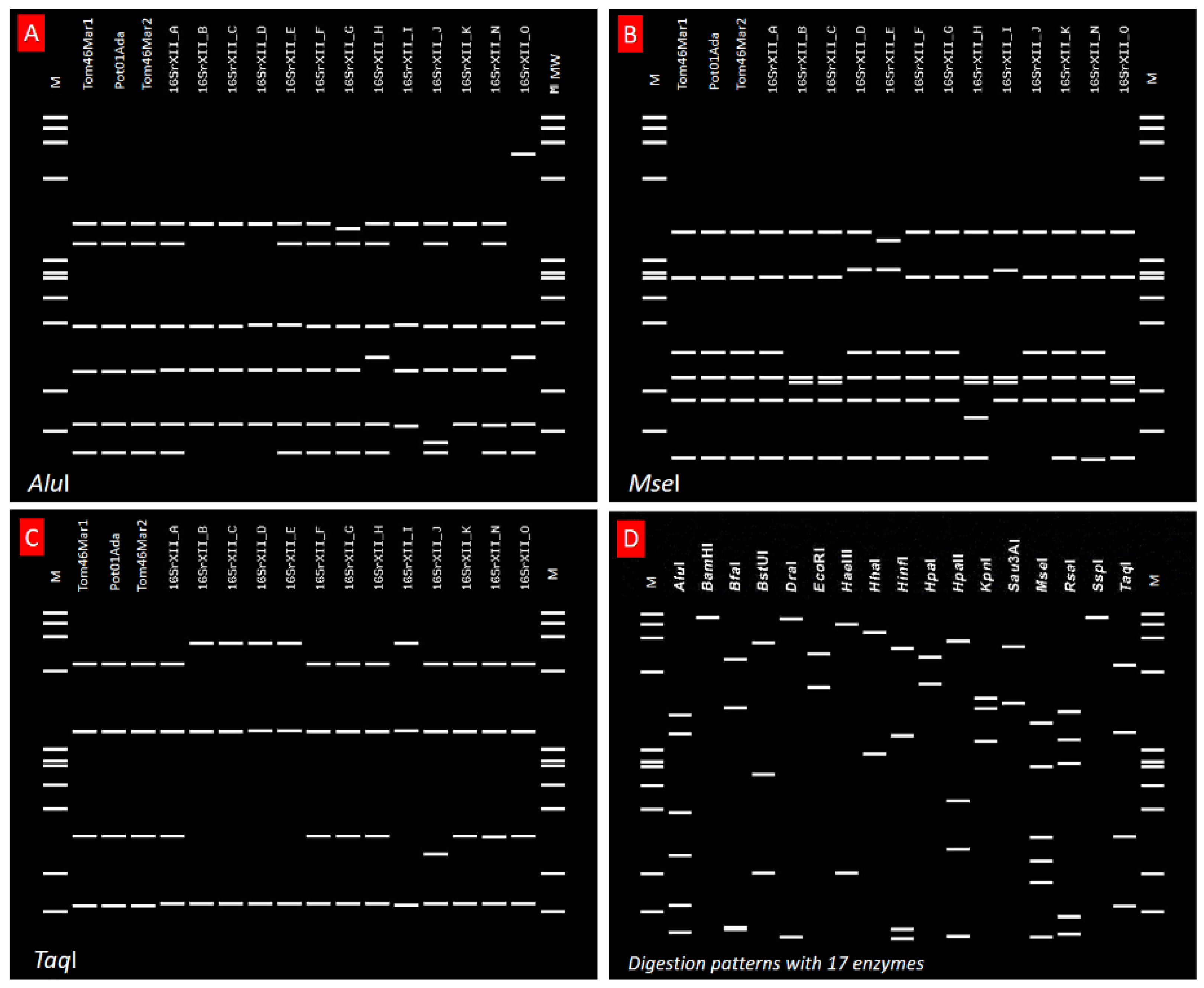
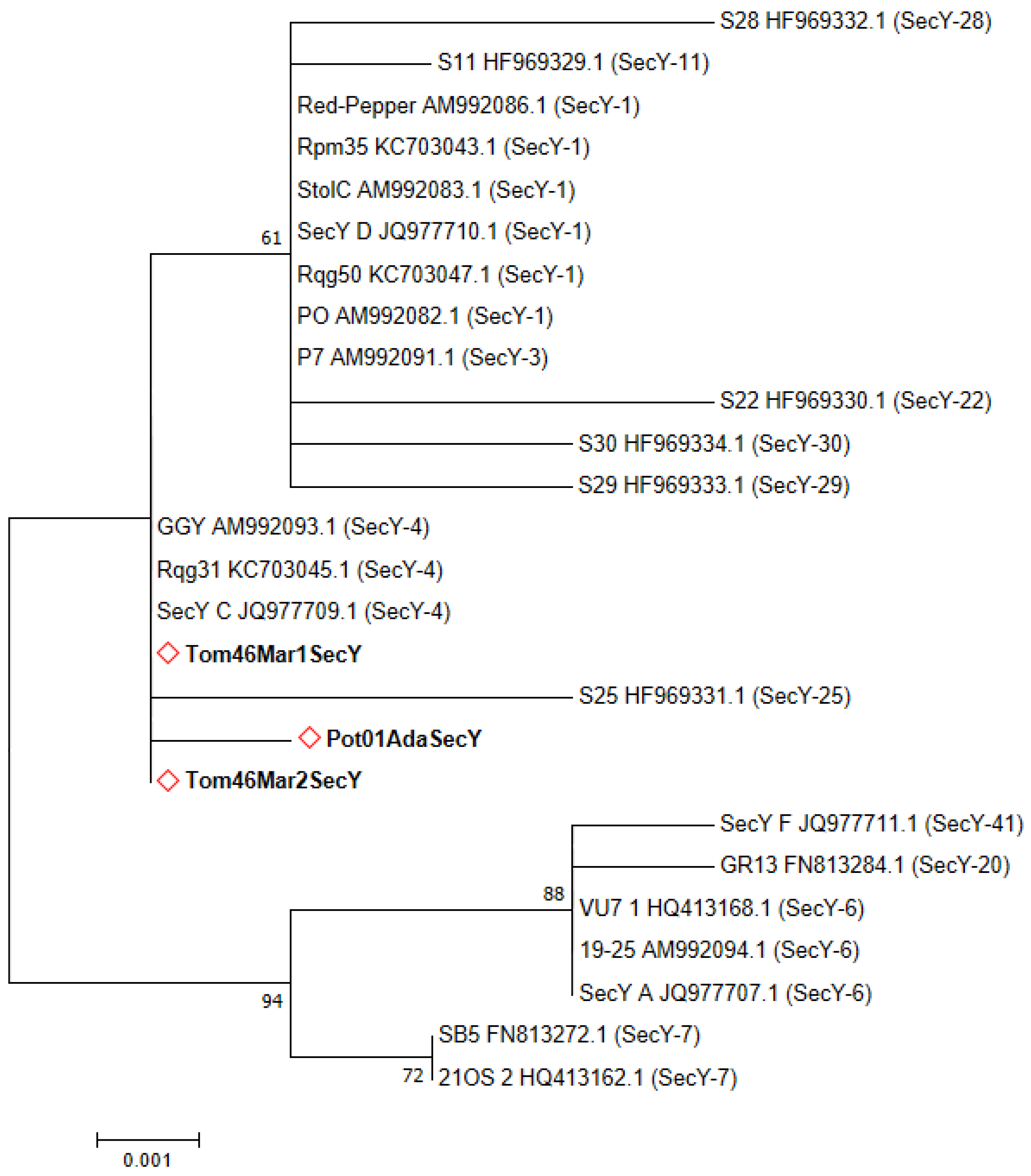
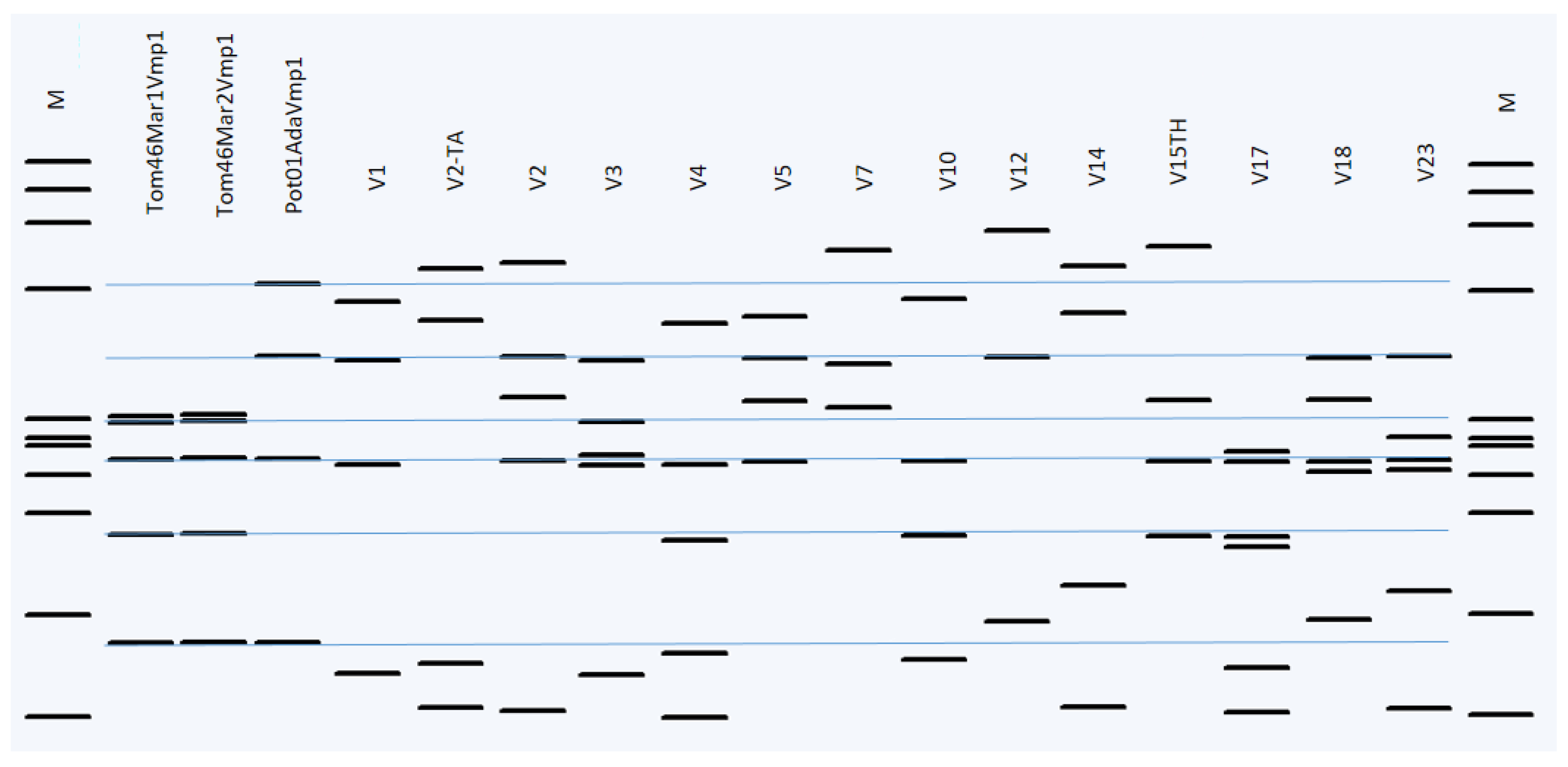
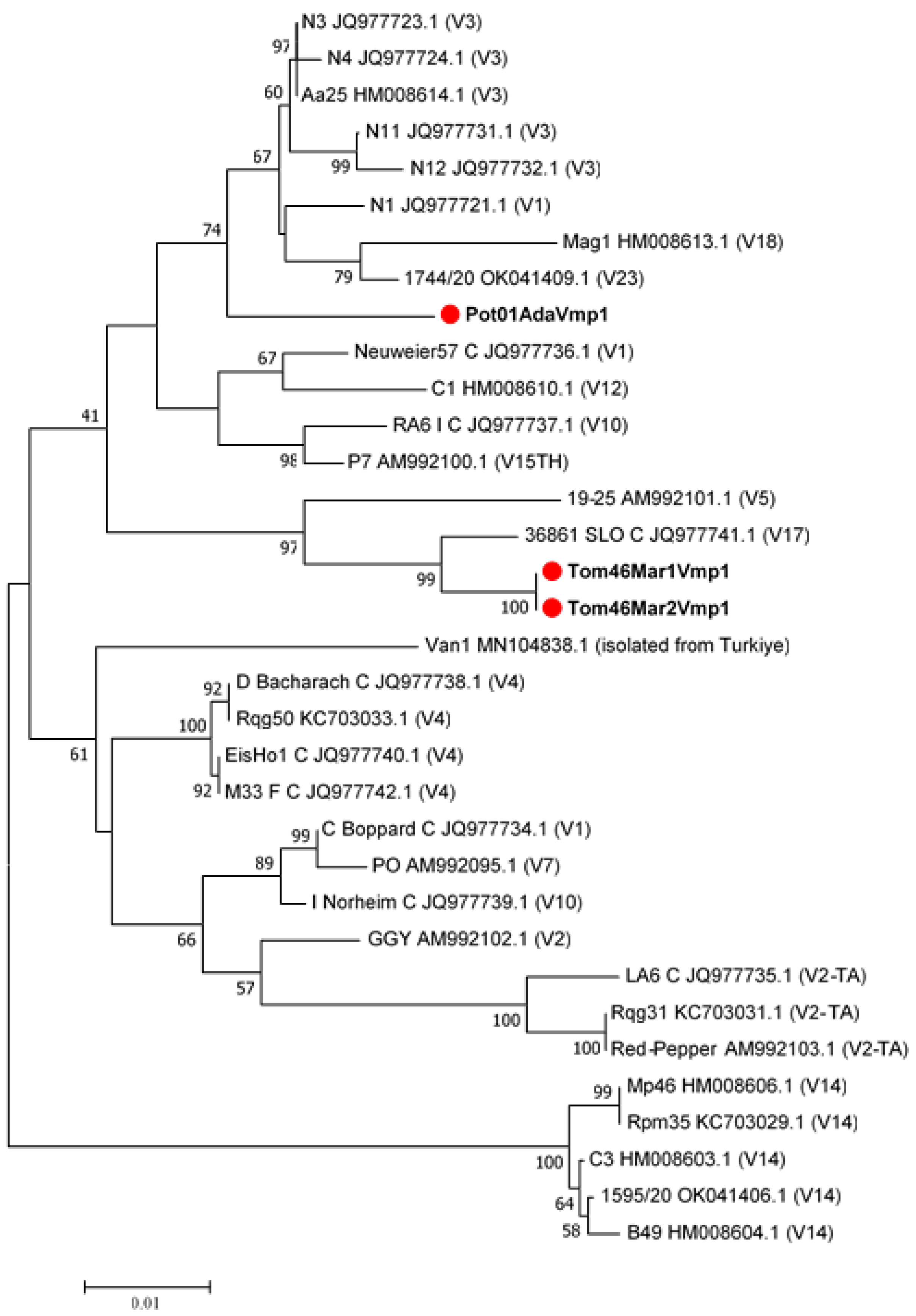
2.2. Sequence Typing by RFLP Analyses and Phytogenetic Relationships
3. Discussion
4. Material and Methods
4.1. Sources of Nucleic Acids and Field Sampling
4.2. Isolation of Nucleic Acids and PCR Detection of Phytoplasmas
4.3. Molecular Characterization of the Strains Based on Tuf, VMP1, and SecY Genes
4.4. Sequencing and Phylogenetic Analyses
4.5. In Silico RFLP Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- TUIK Turkish Statistical Institute (TUIK). Crop Production Statistics. 2021. Available online: https://data.tuik.gov.tr/ (accessed on 7 September 2022).
- Sertkaya, G.; Martini, M.; Musetti, R.; Osler, R. Detection and molecular characterization of phytoplasmas infecting sesame and solanaceous crops in Turkey. Bull. Insectology 2007, 60, 141. [Google Scholar]
- Özdemir, N.; Saygili, H.; Sahin, F.; Karsavuran, Y.; Bayrak, O.F.; Oral, B. Host range and genetic characterization of a phytoplasma causing tomato stolbur disease in Turkey. Acta Hortic. 2009, 808, 255–262. [Google Scholar] [CrossRef]
- Eroglu, S.; Ozbek, H.; Sahin, F. First Report of Group 16SrXII Phytoplasma causing stolbur disease in potato plants in the eastern and southern anatolia regions of Turkey. Plant. Dis. 2010, 94, 1374. [Google Scholar] [CrossRef] [PubMed]
- Çağlar, B.K.; Elbeaino, T.; Pehlivan, D.; Fidan, H.; Baloğlu, S. Genetically comparing of stolbur phytoplasma on potato plants from kayseri and sivas provinces in Turkey. In Proceedings of the 15th Triennial Meeting of the Virology Section of the European Association of Potato Research (EAPR), Antalya, Turkey, 23–31 May 2013; p. 34. [Google Scholar]
- Şimşek, E.; Güldür, M. Detection and molecular characterization of phytoplasmas based on 16s rdna gene region by phylogenetic and in silico rflp analysis of local grapevine cultivars in Şanlıurfa and Adıyaman. Harran Tarım Ve Gıda Bilim. Derg. 2021, 25, 204–213. [Google Scholar] [CrossRef]
- Çağlar, B.K.; Şimşek, E.; Dikilitas, M.; Bertaccini, A. Characterization of ‘Candidatus Phytoplasma Solani’ associated with a maize leaf reddening disease in Turkey. J. Phytopathol. 2021, 169, 658–666. [Google Scholar] [CrossRef]
- Gazel, M.; Çağlayan, K.; Başpınar, H.; Mejia, J.F.; Paltrinieri, S.; Bertaccini, A.; Contaldo, N. Detection and identification of phytoplasmas in pomegranate trees with yellows symptoms. J. Phytopathol. 2016, 164, 136–140. [Google Scholar] [CrossRef]
- Usta, M.; Güller, A.; Sipahioğlu, H.M. Detection and characterization of two phytoplasma lineages on cucumber (Cucumis sativus L.) with same symptomatology based on virtual rflp and nucleotide sequence analysis of 16S RDNA. Yuz. Yil Univ. J. Agric. Sci. 2017, 27, 299–308. [Google Scholar]
- Jović, J.; Cvrković, T.; Mitrović, M.; Krnjajić, S.; Redinbaugh, M.G.; Pratt, R.C.; Gingery, R.E.; Hogenhout, S.A.; Toševski, I. Roles of stolbur phytoplasma and Reptalus panzeri (Cixiinae, Auchenorrhyncha) in the epidemiology of maize redness in Serbia. Eur. J. Plant. Pathol. 2007, 118, 85–89. [Google Scholar] [CrossRef]
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. “Candidatus Phytoplasma Solani”, a novel taxon associated with stolbur- and bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef]
- Sforza, R.; Clair, D.; Daire, X.; Larrue, J.; Boudon-Padieu, E. The role of Hyalesthes obsoletus (Hemiptera: Cixiidae) in the occurrence of bois noir of grapevines in France. J. Phytopathol. 1998, 146, 549–556. [Google Scholar] [CrossRef]
- Maixner, M. Transmission of German grapevine yellows (Vergilbungskrankheit) by the planthopper Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Vitis 1994, 33, 103–104. [Google Scholar]
- Pierro, R.; Passera, A.; Panattoni, A.; Rizzo, D.; Stefani, L.; Bartolini, L.; Casati, P.; Luvisi, A.; Quaglino, F.; Materazzi, A. Prevalence of a ‘Candidatus Phytoplasma Solani’ Strain, so far associated only with other hosts, in bois noir-affected grapevines within tuscan vineyards. Ann. Appl. Biol. 2018, 173, 202–212. [Google Scholar] [CrossRef]
- Mitrović, M.; Jakovljević, M.; Jović, J.; Krstić, O.; Kosovac, A.; Trivellone, V.; Jermini, M.; Toševski, I.; Cvrković, T. ‘Candidatus Phytoplasma Solani’ genotypes associated with potato stolbur in serbia and the role of Hyalesthes obsoletus and Reptalus panzeri (hemiptera, cixiidae) as natural vectors. Eur. J. Plant. Pathol. 2016, 144, 619–630. [Google Scholar] [CrossRef]
- Nair, S.; Manimekalai, R. Phytoplasma diseases of plants: Molecular diagnostics and way forward. World J. Microbiol. Biotechnol. 2021, 37, 102. [Google Scholar] [CrossRef]
- Ray, M.R.; Sinha, D. Phytoplasma. In Methods in Molecular Biology; Dickinson, M., Hodgetts, J., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 938. [Google Scholar]
- Fabre, A.; Danet, J.L.; Foissac, X. The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene 2011, 472, 37–41. [Google Scholar] [CrossRef]
- Passera, A.; Zhao, Y.; Murolo, S.; Pierro, R.; Arsov, E.; Mori, N.; Moussa, A.; Silletti, M.R.; Casati, P.; Panattoni, A.; et al. Multilocus genotyping reveals new molecular markers for differentiating distinct genetic lineages among “Candidatus Phytoplasma Solani” strains associated with grapevine bois noir. Pathogens 2020, 9, 970. [Google Scholar] [CrossRef]
- Pierro, R.; Panattoni, A.; Passera, A.; Materazzi, A.; Luvisi, A.; Loni, A.; Ginanni, M.; Lucchi, A.; Bianco, P.A.; Quaglino, F. Proposal of a new bois noir epidemiological pattern related to ‘Candidatus phytoplasma solani’ strains characterized by a possible moderate virulence in Tuscany. Pathogens 2020, 9, 268. [Google Scholar] [CrossRef]
- Cvrković, T.; Jović, J.; Mitrović, M.; Krstić, O.; Toševski, I. Experimental and molecular evidence of Reptalus Panzeri as a natural vector of bois noir. Plant. Pathol. 2014, 63, 42–53. [Google Scholar] [CrossRef]
- Quaglino, F.; Sanna, F.; Moussa, A.; Faccincani, M.; Passera, A.; Casati, P.; Bianco, P.A.; Mori, N. identification and ecology of alternative insect vectors of ‘Candidatus Phytoplasma Solani’ to grapevine. Sci. Rep. 2019, 9, 19522. [Google Scholar] [CrossRef]
- Pierro, R.; De Pascali, M.; Panattoni, A.; Passera, A.; Materazzi, A.; De Bellis, L.; Luvisi, A.; Bianco, P.A.; Quaglino, F. In silico three-dimensional (3d) modeling of the secy protein of ‘Candidatus Phytoplasma Solani’ strains associated with grapevine “bois noir” and its possible relationship with strain virulence. Int. J. Plant Biol. 2022, 13, 15–30. [Google Scholar] [CrossRef]
- Aryan, A.; Brader, G.; Mörtel, J.; Pastar, M.; Riedle-Bauer, M. An abundant “Candidatus Phytoplasma Solani” Tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur. J. Plant. Pathol. 2014, 140, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Johannesen, J.; Foissac, X.; Kehrli, P.; Maixner, M. Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS ONE 2012, 7, e51809. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, E.; Murolo, S.; Ravari, S.B.; Salehi, M.; Romanazzi, G. Molecular typing of ‘ Candidatus Phytoplasma Solani’ in Iranian vineyards. Plant. Dis. 2019, 103, 2412–2416. [Google Scholar] [CrossRef] [PubMed]
- Plavec, J.; Križanac, I.; Budinšćak, Ž.; Škorić, D.; Musić, M.Š. A case study of fd and bn phytoplasma variability in croatia: Multigene sequence analysis approach. Eur. J. Plant. Pathol. 2015, 142, 591–601. [Google Scholar] [CrossRef]
- Kosovac, A.; Radonjić, S.; Hrnčić, S.; Krstić, O.; Toševski, I.; Jović, J. Molecular tracing of the transmission routes of bois noir in Mediterranean vineyards of Montenegro and experimental evidence for the epidemiological role of Vitex agnus-castus (Lamiaceae) and associated Hyalesthes obsoletus (Cixiidae). Plant. Pathol. 2016, 65, 285–298. [Google Scholar] [CrossRef]
- Atanasova, B.; Jakovljević, M.; Spasov, D.; Jović, J.; Mitrović, M.; Toševski, I.; Cvrković, T. The molecular epidemiology of bois noir grapevine yellows caused by ‘Candidatus Phytoplasma Solani’ in the Republic of Macedonia. Eur. J. Plant. Pathol. 2015, 142, 759–770. [Google Scholar] [CrossRef]
- Mori, H.; Ito, K. The sec protein-translocation pathway. Trends Microbiol. 2001, 9, 494–500. [Google Scholar] [CrossRef]
- Lee, I.M.; Zhao, Y.; Bottner, K.D. SecY gene sequence analysis for finer differentiation of diverse strains in the aster yellows phytoplasma group. Mol. Cell. Probes 2006, 20, 87–91. [Google Scholar] [CrossRef]
- Ma, C.; Wu, X.; Sun, D.; Park, E.; Catipovic, M.A.; Rapoport, T.A.; Gao, N.; Li, L. Structure of the substrate-engaged SecA-SecY protein translocation machine. Nat. Commun. 2019, 10, 2872. [Google Scholar] [CrossRef]
- Tsirigotaki, A.; De Geyter, J.; Šoštarić, N.; Economou, A.; Karamanou, S. Protein export through the bacterial sec pathway. Nat. Rev. Microbiol. 2017, 15, 21–36. [Google Scholar] [CrossRef]
- Quaglino, F.; Maghradze, D.; Casati, P.; Chkhaidze, N.; Lobjanidze, M.; Ravasio, A.; Passera, A.; Venturini, G.; Failla, O.; Bianco, P.A. Identification and characterization of new ‘Candidatus Phytoplasma Solani’ strains associated with bois noir disease in Vitis Vinifera, L. Cultivars Showing a Range of Symptom Severity in Georgia, the Caucasus Region. Plant. Dis. 2016, 100, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Murolo, S.; Romanazzi, G. In-vineyard population structure of ‘Candidatus Phytoplasma Solani’ using multilocus sequence typing analysis. Infect. Genet. Evol. 2015, 31, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Cimerman, A.; Pacifico, D.; Salar, P.; Marzachi, C.; Foissac, X. Striking diversity of vmp1, a variable gene encoding a putative membrane protein of the stolbur phytoplasma. Appl. Environ. Microbiol. 2009, 75, 2951–2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, R.E.; Zhao, Y.; Dally, E.L.; Lee, I.M.; Jomantiene, R.; Douglas, S.M. “Candidatus Phytoplasma Pruni”, a novel taxon associated with x-disease of stone fruits, Prunus spp.: Multilocus characterization based on 16S RRNA, SecY, and ribosomal protein genes. Int. J. Syst. Evol. Microbiol. 2013, 63, 766–776. [Google Scholar] [CrossRef]
- Langer, M.; Maixner, M. Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur-group based on rflp-analysis of non-ribosomal DNA. Vitis 2004, 43, 191–199. [Google Scholar]
- Foissac, X.; Carle, P.; Fabre, A.; Salar, P.; Danet, J.-L.; Stolbur Euromed Consortium. ‘Candidatus Phytoplasma Solani’ Genome Project and Genetic Diversity in the Euro-Mediterranean Basin Invited Cconference. In Proceedings of the Third European Bois Noir Workshop, Barcelona, Spain, 20–21 March 2013. [Google Scholar]
- Nogay, A.; Ternar, Ş.; Ünal, E. Marmara bölgesi’nde domateslerde görülen stolbur hastaliği üzerinde araştirmalar. Bitki Koruma Bülteni 1988, 28, 79–98. [Google Scholar]
- Güller, A.; Usta, M. Stolbur and clover proliferation phytoplasma infections in tomato from Bingöl Province, Turkey. Türk Tarım Ve Doğa Bilim. Derg. 2020, 7, 855–866. [Google Scholar] [CrossRef]
- Çağlar, B.K.; Elbeaino, T.; Küsek, M.; Pehlivan, D.; Fidan, H.; Portakaldalı, M. Stolbur phytoplasma infections in potato and tomato plants from different locations in Turkey. J. Turk. Phytopathol. 2010, 39, 1–8. [Google Scholar]
- Sharon, R.; Soroker, V.; Wesley, S.D.; Zahavi, T.; Harari, A.; Weintraub, P.G. Vitex agnus-castus is a preferred host plant for Hyalesthes obsoletus. J. Chem. Ecol. 2005, 31, 1051–1063. [Google Scholar] [CrossRef]
- Bayram, S.; Zeybekoglu, U.; Soylemezoglu, G.; Canik, D.; Karavin, M.; Cakir, A.; Ertunc, F. Presence of putative insect vectors of grapevine yellows phytoplasmas in Turkey. Phytopathog. Mollicutes 2014, 4, 22–26. [Google Scholar] [CrossRef]
- Pala, F. Observation of weed species, frequency and density in common barley (Hordeum vulgare L.) fields of diyarbakir, Turkey: A case study. Tarım Bilim. Derg. 2020, 26, 164–172. [Google Scholar] [CrossRef]
- Usta, M.; Güller, A.; Demirel, S. Molecular identification of ‘candidatus phytoplasma solani’ using secy and vmp1 genes in tomato plants from van province. Yuz. Yil Univ. J. Agric. Sci. 2021, 31, 951–960. [Google Scholar] [CrossRef]
- Arricau-Bouvery, N.; Duret, S.; Dubrana, M.P.; Batailler, B.; Desqué, D.; Béven, L.; Danet, J.L.; Monticone, M.; Bosco, D.; Malembic-Maher, S.; et al. Variable membrane protein a of flavescence dorée phytoplasma binds the midgut perimicrovillar membrane of Euscelidius variegatus and promotes adhesion to its epithelial cells. Appl. Environ. Microbiol. 2018, 84, e02487-17. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, U.; Seemüller, E. Detection of DNA of plant pathogenic mycoplasmalike organisms by a polymerase chain reaction that amplifies a sequence of the 16S RRNA gene. Phytopathology 1992, 82, 828–832. [Google Scholar] [CrossRef]
- Deng, S.; Hiruki, C. Amplification of 16S RRNA genes from culturable and nonculturable mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Smart, C.D.; Schneider, B.; Blomquist, C.L.; Guerra, L.J.; Harrison, N.A.; Ahrens, U.; Lorenz, K.H.; Seemüller, E.; Kirkpatrick, B.C. Phytoplasma-specific PCR primers based on sequences of the 16S-23S RRNA spacer region. Appl. Environ. Microbiol. 1996, 62, 2988–2993. [Google Scholar] [CrossRef] [Green Version]
- Gundersen, D.; Lee, I. Ultrasensitive detection of phytoplasmas by nested-pcr assays using two universal primer pairs. Phytopathol. Mediterr. 1996, 35, 144–151. [Google Scholar]
- Fialová, R.; Válová, P.; Balakishiyeva, G.; Danet, J.L.; Šafárová, D.; Foissac, X.; Navrátil, M. Genetic variability of “stolbur” phytoplasma in annual crop and wild plant species in South Moravia. J. Plant. Pathol. 2009, 91, 411–416. [Google Scholar]
- Schneider, B.; Gibb, K.S.; Seemüller, E. Sequence and RFLP Analysis of the elongation factor tu gene used in differentiation and classification of phytoplasmas. Microbiology 1997, 143, 3381–3389. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wei, W.; Lee, I.-M.; Shao, J.; Suo, X.; Davis, R.E. construction of an interactive online phytoplasma classification tool, iphyclassifier, and its application in analysis of the peach x-disease phytoplasma group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59, 2582–2593. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bartoszyk, I.M. Revised classification scheme of phytoplasmas based on rflp analyses of 16S RRNA and ribosomal protein gene sequences. Int. J. Syst. Bacteriol. 1998, 48, 1153–1169. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Çağlar, B.K.; Şimşek, E. Detection and Multigene Typing of ‘Candidatus Phytoplasma solani’-Related Strains Infecting Tomato and Potato Plants in Different Regions of Turkey. Pathogens 2022, 11, 1031. https://doi.org/10.3390/pathogens11091031
Çağlar BK, Şimşek E. Detection and Multigene Typing of ‘Candidatus Phytoplasma solani’-Related Strains Infecting Tomato and Potato Plants in Different Regions of Turkey. Pathogens. 2022; 11(9):1031. https://doi.org/10.3390/pathogens11091031
Chicago/Turabian StyleÇağlar, Behçet Kemal, and Eray Şimşek. 2022. "Detection and Multigene Typing of ‘Candidatus Phytoplasma solani’-Related Strains Infecting Tomato and Potato Plants in Different Regions of Turkey" Pathogens 11, no. 9: 1031. https://doi.org/10.3390/pathogens11091031
APA StyleÇağlar, B. K., & Şimşek, E. (2022). Detection and Multigene Typing of ‘Candidatus Phytoplasma solani’-Related Strains Infecting Tomato and Potato Plants in Different Regions of Turkey. Pathogens, 11(9), 1031. https://doi.org/10.3390/pathogens11091031





