Genomic Islands Shape the Genetic Background of Both JP2 and Non-JP2 Aggregatibacter actinomycetemcomitans
Abstract
:1. Introduction
2. Results and Discussion
2.1. Features of A. actinomycetemcomitans Genomes
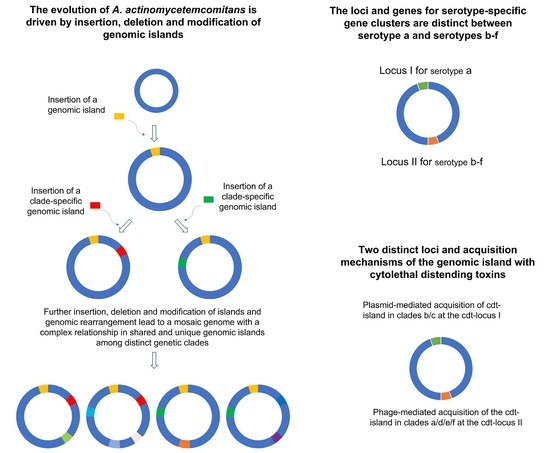
2.2. Islands May Drive the Evolutionary Divergence of the Species: Distribution of the Homologous Genomic Island Groups among the A. actinomycetemcomitans Strains
2.3. Acquisition of Leukotoxin Operon Marked the Distinction between A. actinomycetemcomitans and A. aphrophilus
2.4. Cytolethal Distending Toxin Operon Was Independently Acquired in Different A. actinomycetemcomitans Strains
2.5. Serotype-Specific Gene Clusters Are Found in Two Different Loci
3. Materials and Methods
3.1. Identification of Core Genes, Accessory Genes, and Genomic Islands
3.2. Grouping of Genomic Islands
3.3. Features of Island Insertion Sites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haubek, D. The highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans: Evolutionary aspects, epidemiology and etiological role in aggressive periodontitis. APMIS Suppl. 2010, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Brogan, J.M.; Lally, E.T.; Poulsen, K.; Kilian, M.; Demuth, D.R. Regulation of Actinobacillus actinomycetemcomitans leukotoxin expression: Analysis of the promoter regions of leukotoxic and minimally leukotoxic strains. Infect. Immun. 1994, 62, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Kittichotirat, W.; Bumgarner, R.E.; Chen, C. Evolutionary Divergence of Aggregatibacter actinomycetemcomitans. J. Dent. Res. 2016, 95, 94–101. [Google Scholar] [CrossRef]
- Kittichotirat, W.; Bumgarner, R.E.; Asikainen, S.; Chen, C. Identification of the pangenome and its components in 14 distinct Aggregatibacter actinomycetemcomitans strains by comparative genomic analysis. PLoS ONE 2011, 6, e22420. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Kittichotirat, W.; Mayer, M.P.; Hall, R.; Bumgarner, R.; Chen, C. Comparative genomic hybridization and transcriptome analysis with a pan-genome microarray reveal distinctions between JP2 and non-JP2 genotypes of Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 2013, 28, 1–17. [Google Scholar] [CrossRef]
- Karched, M.; Furgang, D.; Planet, P.J.; DeSalle, R.; Fine, D.H. Genome sequence of Aggregatibacter actinomycetemcomitans RHAA1, isolated from a rhesus macaque, an Old World primate. J. Bacteriol 2012, 194, 1275–1276. [Google Scholar] [CrossRef]
- Fujise, O.; Lakio, L.; Wang, Y.; Asikainen, S.; Chen, C. Clonal distribution of natural competence in Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol 2004, 19, 340–342. [Google Scholar] [CrossRef]
- Wang, Y.; Goodman, S.D.; Redfield, R.J.; Chen, C. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J. Bacteriol. 2002, 184, 3442–3449. [Google Scholar] [CrossRef]
- Jorth, P.; Whiteley, M. An evolutionary link between natural transformation and CRISPR adaptive immunity. MBio 2012, 3, e00309-12. [Google Scholar] [CrossRef]
- Nedergaard, S.; Kobel, C.M.; Nielsen, M.B.; Moller, R.T.; Jensen, A.B.; Norskov-Lauritsen, N. Whole genome sequencing of Aggregatibacter actinomycetemcomitans cultured from blood stream infections reveals three major phylogenetic groups including a novel lineage expressing serotype a membrane o polysaccharide. Pathogens 2019, 8, 256. [Google Scholar] [CrossRef] [Green Version]
- van der Reijden, W.A.; Brunner, J.; Bosch-Tijhof, C.J.; van Trappen, S.; Rijnsburger, M.C.; de Graaff, M.P.; van Winkelhoff, A.J.; Cleenwerck, I.; de Vos, P. Phylogenetic variation of Aggregatibacter actinomycetemcomitans serotype e reveals an aberrant distinct evolutionary stable lineage. Infect. Genet. Evol. 2010, 10, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Murra, M.; Lutzen, L.; Barut, A.; Zbinden, R.; Lund, M.; Villesen, P.; Norskov-Lauritsen, N. Whole-Genome Sequencing of Aggregatibacter Species Isolated from Human Clinical Specimens and Description of Aggregatibacter kilianii sp. nov. J. Clin. Microbiol 2018, 56, e00053-18. [Google Scholar] [CrossRef] [PubMed]
- Tempro, P.J.; Slots, J. Selective medium for the isolation of Haemophilus aphrophilus from the human periodontium and other oral sites and the low proportion of the organism in the oral flora. J. Clin. Microbiol 1986, 23, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Asikainen, S.; Chen, C.; Saarela, M.; Saxen, L.; Slots, J. Clonal specificity of Actinobacillus actinomycetemcomitans in destructive periodontal disease. Clin. Infect. Dis. 1997, 25 (Suppl. 2), S227–S229. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Kaplan, J.B.; Kachlany, S.C.; Schreiner, H.C. How we got attached to Actinobacillus actinomycetemcomitans: A model for infectious diseases. Periodontology 2000 2006, 42, 114–157. [Google Scholar] [CrossRef]
- Aberg, C.H.; Kelk, P.; Johansson, A. Aggregatibacter actinomycetemcomitans: Virulence of its leukotoxin and association with aggressive periodontitis. Virulence 2015, 6, 188–195. [Google Scholar] [CrossRef]
- Kilian, M.; Frandsen, E.V.; Haubek, D.; Poulsen, K. The etiology of periodontal disease revisited by population genetic analysis. Periodontology 2000 2006, 42, 158–179. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bueno, L.C.; Hansen, E.J.; DiRienzo, J.M. Identification of a cytolethal distending toxin gene locus and features of a virulence-associated region in Actinobacillus actinomycetemcomitans. Infect. Immun. 1999, 67, 1227–1237. [Google Scholar] [CrossRef]
- Shenker, B.J.; McKay, T.; Datar, S.; Miller, M.; Chowhan, R.; Demuth, D. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2 arrest in human T cells. J. Immunol. 1999, 162, 4773–4780. [Google Scholar]
- Doungudomdacha, S.; Volgina, A.; DiRienzo, J.M. Evidence that the cytolethal distending toxin locus was once part of a genomic island in the periodontal pathogen Aggregatibacter (Actinobacillus) actinomycetemcomitans strain Y4. J. Med. Microbiol. 2007, 56, 1519–1527. [Google Scholar] [CrossRef]
- Asikainen, S.; Chen, C.; Slots, J. Actinobacillus actinomycetemcomitans genotypes in relation to serotypes and periodontal status. Oral Microbiol Immunol. 1995, 10, 65–68. [Google Scholar] [CrossRef]
- Dogan, B.; Saarela, M.H.; Jousimies-Somer, H.; Alaluusua, S.; Asikainen, S. Actinobacillus actinomycetemcomitans serotype e--biotypes, genetic diversity and distribution in relation to periodontal status. Oral Microbiol Immunol. 1999, 14, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.W.; Asikainen, S.; Dogan, B.; Suda, R.; Lai, C.H. Relationship of Actinobacillus actinomycetemcomitans serotype b to aggressive periodontitis: Frequency in pure cultured isolates. J. Periodontol. 2004, 75, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Asikainen, S.; Lai, C.H.; Alaluusua, S.; Slots, J. Distribution of Actinobacillus actinomycetemcomitans serotypes in periodontal health and disease. Oral Microbiol Immunol. 1991, 6, 115–118. [Google Scholar] [CrossRef]
- Kaplan, J.B.; Perry, M.B.; MacLean, L.L.; Furgang, D.; Wilson, M.E.; Fine, D.H. Structural and genetic analyses of O polysaccharide from Actinobacillus actinomycetemcomitans serotype f. Infect. Immun. 2001, 69, 5375–5384. [Google Scholar] [CrossRef]
- Perry, M.B.; MacLean, L.L.; Gmur, R.; Wilson, M.E. Characterization of the O-polysaccharide structure of lipopolysaccharide from Actinobacillus actinomycetemcomitans serotype b. Infect. Immun. 1996, 64, 1215–1219. [Google Scholar] [CrossRef]
- Perry, M.B.; MacLean, L.M.; Brisson, J.R.; Wilson, M.E. Structures of the antigenic O-polysaccharides of lipopolysaccharides produced by Actinobacillus actinomycetemcomitans serotypes a, c, d and e. Eur. J. Biochem. 1996, 242, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E.; Schifferle, R.E. Evidence that the serotype b antigenic determinant of Actinobacillus actinomycetemcomitans Y4 resides in the polysaccharide moiety of lipopolysaccharide. Infect. Immun. 1991, 59, 1544–1551. [Google Scholar] [CrossRef]
- Shibuya, N.; Amano, K.; Azuma, J.; Nishihara, T.; Kitamura, Y.; Noguchi, T.; Koga, T. 6-Deoxy-D-talan and 6-deoxy-L-talan. Novel serotype-specific polysaccharide antigens from Actinobacillus actinomycetemcomitans. J. Biol Chem. 1991, 266, 16318–16323. [Google Scholar] [CrossRef]
- Suzuki, N.; Nakano, Y.; Yoshida, Y.; Nezu, T.; Terada, Y.; Yamashita, Y.; Koga, T. Guanosine diphosphate-4-keto-6-deoxy-d-mannose reductase in the pathway for the synthesis of GDP-6-deoxy-d-talose in Actinobacillus actinomycetemcomitans. Eur. J. Biochem. 2002, 269, 5963–5971. [Google Scholar] [CrossRef]
- Suzuki, N.; Nakano, Y.; Yoshida, Y.; Nakao, H.; Yamashita, Y.; Koga, T. Genetic analysis of the gene cluster for the synthesis of serotype a-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys Acta. 2000, 1517, 135–138. [Google Scholar] [CrossRef]
- Nakano, Y.; Yoshida, Y.; Yamashita, Y.; Koga, T. A gene cluster for 6-deoxy-L-talan synthesis in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta. 1998, 1442, 409–414. [Google Scholar] [CrossRef]
- Nakano, Y.; Yoshida, Y.; Suzuki, N.; Yamashita, Y.; Koga, T. A gene cluster for the synthesis of serotype d-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta. 2000, 1493, 259–263. [Google Scholar] [CrossRef]
- Yoshida, Y.; Nakano, Y.; Suzuki, N.; Nakao, H.; Yamashita, Y.; Koga, T. Genetic analysis of the gene cluster responsible for synthesis of serotype e-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta. 1999, 1489, 457–461. [Google Scholar] [CrossRef]
- Yoshida, Y.; Nakano, Y.; Yamashita, Y.; Koga, T. Identification of a genetic locus essential for serotype b-specific antigen synthesis in Actinobacillus actinomycetemcomitans. Infect. Immun. 1998, 66, 107–114. [Google Scholar] [CrossRef]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. RNA-Seq analysis in MeV. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kittichotirat, W.; Bumgarner, R.E.; Chen, C. Genomic Islands Shape the Genetic Background of Both JP2 and Non-JP2 Aggregatibacter actinomycetemcomitans. Pathogens 2022, 11, 1037. https://doi.org/10.3390/pathogens11091037
Kittichotirat W, Bumgarner RE, Chen C. Genomic Islands Shape the Genetic Background of Both JP2 and Non-JP2 Aggregatibacter actinomycetemcomitans. Pathogens. 2022; 11(9):1037. https://doi.org/10.3390/pathogens11091037
Chicago/Turabian StyleKittichotirat, Weerayuth, Roger E. Bumgarner, and Casey Chen. 2022. "Genomic Islands Shape the Genetic Background of Both JP2 and Non-JP2 Aggregatibacter actinomycetemcomitans" Pathogens 11, no. 9: 1037. https://doi.org/10.3390/pathogens11091037
APA StyleKittichotirat, W., Bumgarner, R. E., & Chen, C. (2022). Genomic Islands Shape the Genetic Background of Both JP2 and Non-JP2 Aggregatibacter actinomycetemcomitans. Pathogens, 11(9), 1037. https://doi.org/10.3390/pathogens11091037