Xenophagy as a Strategy for Mycobacterium leprae Elimination during Type 1 or Type 2 Leprosy Reactions: A Systematic Review
Abstract
:1. Introduction
2. Methods
2.1. Study Selection and Eligibility Criteria
2.2. Information Sources and Literature Search Strategies
2.3. Selection and Data Collection
2.4. Data Items
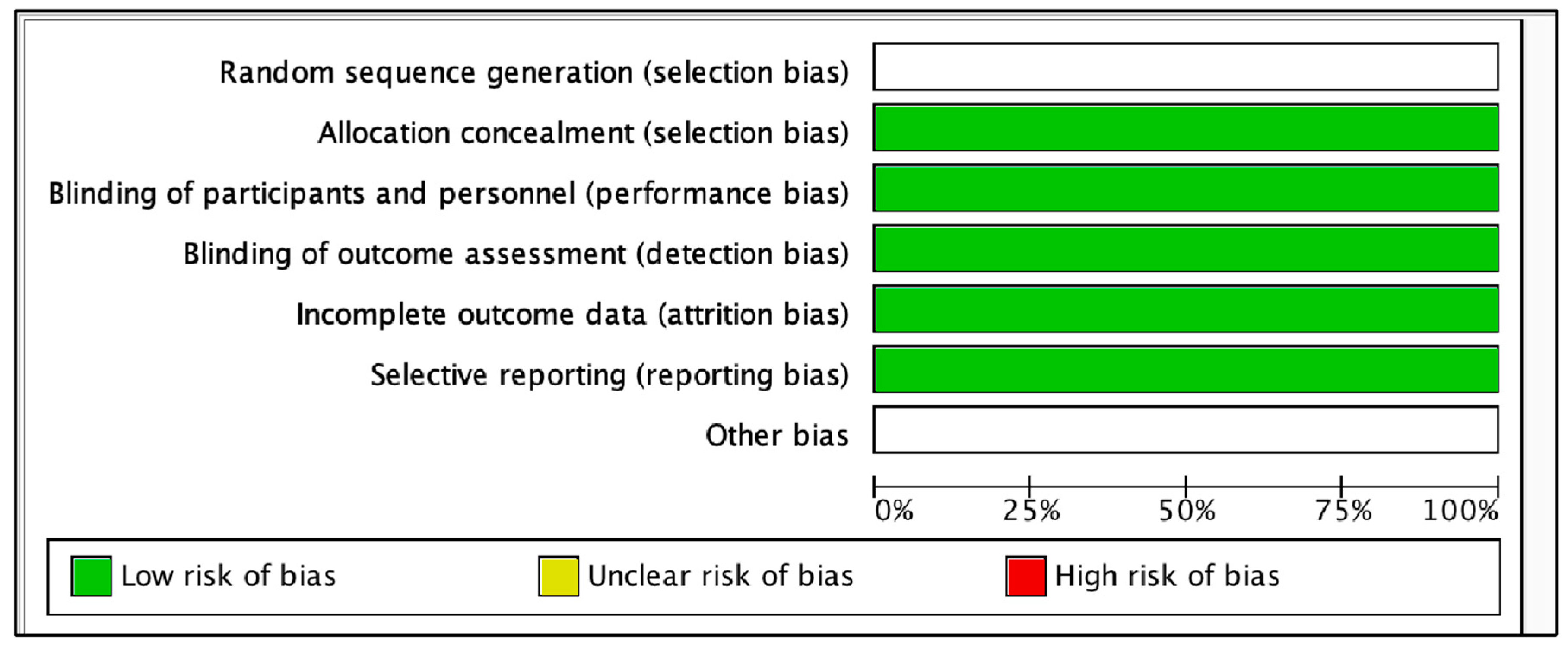
2.5. Risk of Bias Assessment
3. Results
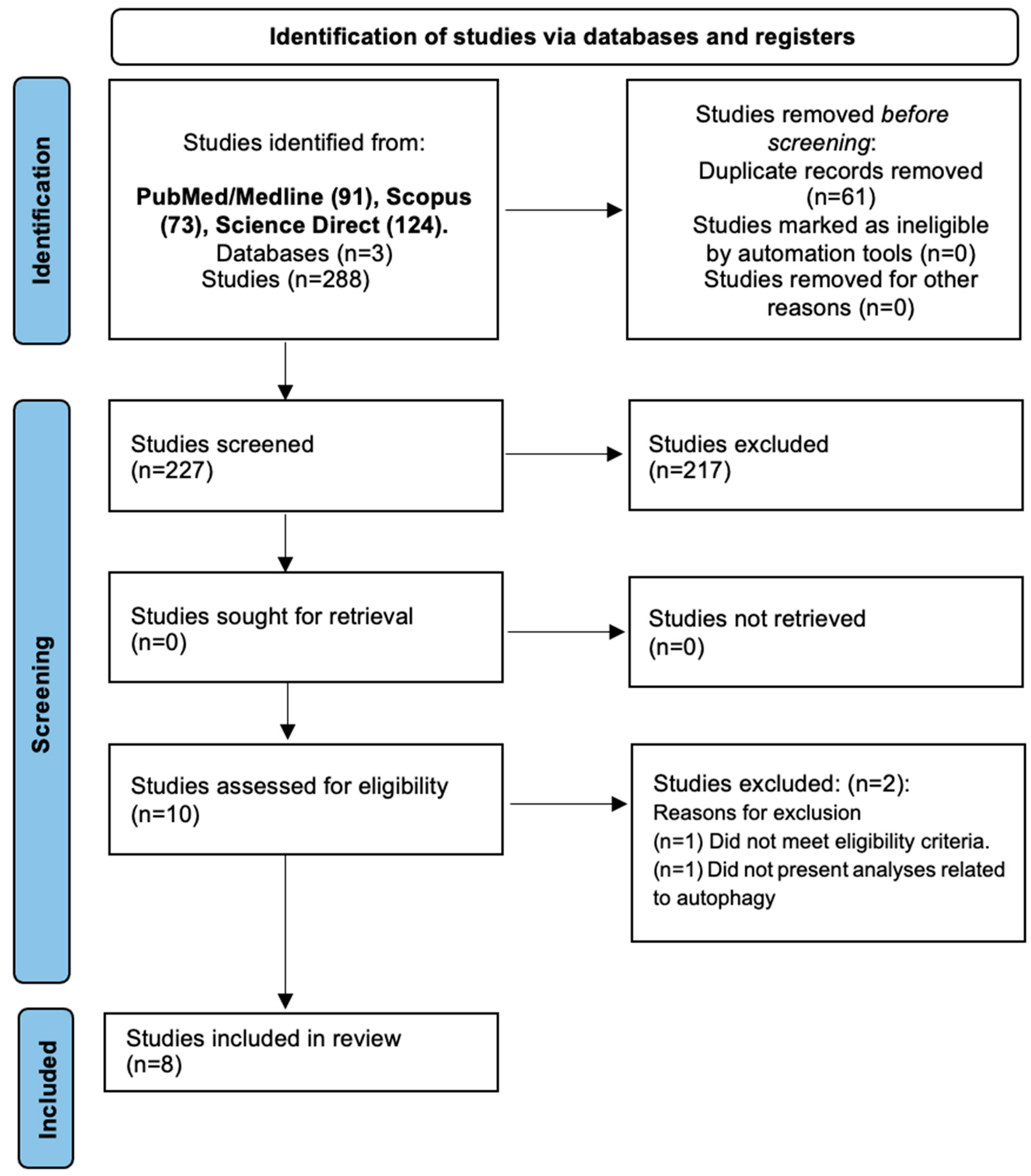
3.1. Search Results
3.2. Study Characteristics
3.2.1. Characteristics of the Studies Included in Humans
3.2.2. In Vitro Studies
3.3. Xenophagy Parameters in Leprosy Patients
3.3.1. Skin Lesion from Leprosy Patients
3.3.2. M. leprae-Stimulated Human Monocytic Cell Line THP-1
3.3.3. Monocyte-Derived Macrophages from Healthy Donors upon Stimulation with M. leprae
4. Discussion
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mungroo, M.R.; Khan, N.A.; Siddiqui, R. Mycobacterium leprae: Pathogenesis, diagnosis, and treatment options. Microb. Pathog. 2020, 149, 104475. [Google Scholar] [CrossRef] [PubMed]
- Mi, Z.; Liu, H.; Zhang, F. Advances in the Immunology and Genetics of Leprosy. Front. Immunol. 2020, 11, 567. [Google Scholar] [CrossRef]
- Abebe, G.; Bonsa, W.K.Z. Treatment Outcomes and Associated Factors in Tuberculosis Patients at Jimma University Medical Center: A 5-Year Retrospective Study Gemeda. Int. J. Mycobacteriology 2017, 6, 239–245. [Google Scholar] [CrossRef]
- Ridley, D.S.; Jopling, W.H. Classification of leprosy according to immunity. A five-group system. Int. J. Lepr. Other Mycobact. Dis. 1966, 34, 255–273. [Google Scholar] [PubMed]
- Silva, B.J.d.A.; Barbosa, M.G.d.M.; Andrade, P.R.; Ferreira, H.; Nery, J.A.d.C.; Côrte-Real, S.; da Silva, G.M.S.; Rosa, P.S.; Fabri, M.; Sarno, E.N.; et al. Autophagy Is an Innate Mechanism Associated with Leprosy Polarization. PLoS Pathog. 2017, 13, 1006103. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, R.O.; de Souza Salles, J.; Sarno, E.N.; Sampaio, E.P. Mycobacterium leprae—Host-cell interactions and genetic determinants in leprosy: An overview. Future Microbiol. 2011, 6, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Eichelmann, K.; González, S.E.G.; Salas-Alanis, J.C.; Ocampo-Candiani, J. Leprosy. An Update: Definition, Pathogenesis, Classification, Diagnosis, and Treatment. Actas Dermo-Sifiliográficas (Engl. Ed.) 2013, 104, 554–563. [Google Scholar] [CrossRef]
- Hooij, A.; Geluk, A. In search of biomarkers for leprosy by unraveling the host immune response to Mycobacterium leprae. Immunol. Rev. 2021, 301, 175–192. [Google Scholar] [CrossRef]
- White, C.; Franco-Paredes, C. Leprosy in the 21st Century. Clin. Microbiol. Rev. 2015, 28, 80–94. [Google Scholar] [CrossRef]
- Froes, L.A.R.; Trindade, M.A.B.; Sotto, M.N. Immunology of leprosy. Int. Rev. Immunol. 2022, 41, 72–83. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Yuk, J.M.; Yoshimori, T.; Jo, E.K. Autophagy and bacterial infectious diseases. Exp. Mol. Med. 2012, 44, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in Human Diseases. N. Engl. J. Med. 2020, 383, 1564–1576. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wei, F.; Li, G. The crosstalk between bacteria and host autophagy: Host defense or bacteria offense. J. Microbiol. 2022, 60, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.J.; Rahimi, N.; Tadi, P. Biochemistry, Ubiquitination; StatPearls: Treasure Island, FL, USA, 2023. [Google Scholar]
- Hu, W.; Chan, H.; Lu, L.; Wong, K.T.; Wong, S.H.; Li, M.X.; Xiao, Z.G.; Cho, C.H.; Gin, T.; Chan, M.T.V.; et al. Autophagy in intracellular bacterial infection. Semin. Cell Dev. Biol. 2020, 101, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, R.O.; Schmitz, V.; Silva, B.J.d.A.; Dias, A.A.; de Souza, B.J.; Barbosa, M.G.d.M.; Esquenazi, D.d.A.; Pessolani, M.C.V.; Sarno, E.N. Innate Immune Responses in Leprosy. Front. Immunol. 2018, 9, 518. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Lu, J.; Shui, T.; Chen, J.; Yang, J.; Yuan, J.; Liu, Y.; Yang, D.; Chen, D.; et al. A negative feedback loop between autophagy and immune responses in Mycobacterium leprae infection. DNA Cell Biol. 2017, 36, 1–9. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Green, S. (Eds.) Cochrane Handbook for Systematic Reviews of Interventions; Wiley: Hoboken, NJ, USA, 2008; p. S38. [Google Scholar] [CrossRef]
- Barbosa, M.G.d.M.; Silva, B.J.d.A.; Assis, T.Q.; Prata, R.B.d.S.; Ferreira, H.; Andrade, P.R.; Oliveira, J.A.d.P.d.; da Silva, G.M.S.; Nery, J.A.d.C.; Sarno, E.N.; et al. Autophagy Impairment Is Associated With Increased Inflammasome Activation and Reversal Reaction Development in Multibacillary Leprosy. Front. Immunol. 2018, 9, 1223. [Google Scholar] [CrossRef]
- de Sousa, J.R.; Falcão, L.F.M.; Virgolino, G.L.; Cruz, M.F.S.; Teixeira, V.F.; Aarão, T.L.d.S.; Furlaneto, I.P.; Carneiro, F.R.O.; Amin, G.; Fuzii, H.T.; et al. Different cell death mechanisms are involved in leprosy pathogenesis. Microb. Pathog. 2022, 166, 105511. [Google Scholar] [CrossRef]
- Silva, B.J.d.A.; Bittencourt, T.L.; Leal-Calvo, T.; Mendes, M.A.; Prata, R.B.d.S.; Barbosa, M.G.d.M.; Andrade, P.R.; Côrte-Real, S.; da Silva, G.M.S.; Moraes, M.O.; et al. Autophagy-associated IL-15 production is involved in the pathogenesis of leprosy type 1 reaction. Cells 2021, 10, 2215. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, J.; Shi, C.; Jing, Z.; Song, N. Autophagy Gene Polymorphism is Associated with Susceptibility to Leprosy by Affecting Inflammatory Cytokines. Inflammation 2014, 37, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, J.; Zhang, L.; Cha, Z.; Han, S.; Shi, W.; Ding, R.; Ma, L.; Xiao, H.; Shi, C.; et al. Mycobacterium leprae Upregulates IRGM Expression in Monocytes and Monocyte-Derived Macrophages. Inflammation 2014, 37, 1028–1034. [Google Scholar] [CrossRef]
- Ma, Y.; Pei, Q.; Zhang, L.; Lu, J.; Shui, T.; Chen, J.; Shi, C.; Yang, J.; Smith, M.; Liu, Y.; et al. Live Mycobacterium leprae inhibits autophagy and apoptosis of infected macrophages and prevents engulfment of host cell by phagocytes. Am. J. Transl. Res. 2018, 10, 2929–2939. [Google Scholar] [PubMed]
- Sugawara-Mikami, M.; Tanigawa, K.; Kawashima, A.; Kiriya, M.; Nakamura, Y.; Fujiwara, Y.; Suzuki, K. Pathogenicity and virulence of Mycobacterium leprae. Virulence 2022, 13, 1985–2011. [Google Scholar] [CrossRef]
- Luo, Y.; Kiriya, M.; Tanigawa, K.; Kawashima, A.; Nakamura, Y.; Ishii, N.; Suzuki, K. Host-Related Laboratory Parameters for Leprosy Reactions. Front. Med. 2021, 8, 694376. [Google Scholar] [CrossRef]
- Kleinnijenhuis, J.; Oosting, M.; Plantinga, T.S.; van der Meer, J.W.; Joosten, L.A.; Crevel, R.V.; Netea, M.G. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response. Immunology 2011, 134, 341–348. [Google Scholar] [CrossRef]
- de Sousa, J.R.; Neto, F.D.L.; Sotto, M.N.; Quaresma, J.A.S. Immunohistochemical characterization of the M4 macrophage population in leprosy skin lesions. BMC Infect. Dis. 2018, 18, 576. [Google Scholar] [CrossRef]
- Strong, E.J.; Lee, S. Targeting Autophagy as a Strategy for Developing New Vaccines and Host-Directed Therapeutics Against Mycobacteria. Front. Microbiol. 2021, 11, 614313. [Google Scholar] [CrossRef]
- Dang, A.T.; Teles, R.M.; Weiss, D.I.; Parvatiyar, K.; Sarno, E.N.; Ochoa, M.T.; Cheng, G.; Gilliet, M.; Bloom, B.R.; Modlin, R.L. IL-26 contributes to host defense against intracellular bacteria. J. Clin. Investig. 2019, 129, 1926–1939. [Google Scholar] [CrossRef]
- Andrade, P.R.; Mehta, M.; Lu, J.; Teles, R.M.B.; Montoya, D.; Scumpia, P.O.; Sarno, E.N.; Ochoa, M.T.; Ma, F.; Pellegrini, M.; et al. The cell fate regulator NUPR1 is induced by Mycobacterium leprae via type I interferon in human leprosy. PLoS Negl. Trop. Dis. 2019, 13, e0007589. [Google Scholar] [CrossRef] [PubMed]
- Montoya, D.; Cruz, D.; Teles, R.M.; Lee, D.J.; Ochoa, M.T.; Krutzik, S.R.; Chun, R.; Schenk, M.; Zhang, X.; Ferguson, B.G.; et al. Divergence of Macrophage Phagocytic and Antimicrobial Programs in Leprosy. Cell Host Microbe 2009, 6, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Rêgo, J.L.; Santana, N.d.L.; Machado, P.R.L.; Ribeiro-Alves, M.; de Toledo-Pinto, T.G.; Castellucci, L.C.; Moraes, M.O. Whole blood profiling of leprosy type 1(reversal) reactions highlights prominence of innate immune response genes. BMC Infect. Dis. 2018, 18, 422. [Google Scholar] [CrossRef] [PubMed]

| Inclusion Criteria | Exclusion Criteria | |
|---|---|---|
| Population | Humans | Animals and other organisms |
| Intervention/Exposure | Leprosy | No leprosy |
| Control | No leprosy patients | - |
| Outcomes | Autophagy parameters | No autophagy parameters |
| Study Design | Clinical studies | Reviews; case reports; letters to editors; comments; etc. |
| Author, Year [Ref.] | Country | Study Design | Group | n | Clinical Form of Leprosy | Sex (Male/Female) | Age, Mean (Range) | BI, Mean (Range) | LBI, Mean (Range) | Leprosy Treatment Status |
|---|---|---|---|---|---|---|---|---|---|---|
| Barbosa et al., 2018 [21] | Brazil | Cohort | WR | 10 | 2 BL/8 LL | 5/5 | 42.9 (25–65) | 4.19 (1.75–5.85) | 4.84 (3.5–5.85) | Pretreatment (24-month follow-up) |
| T1R | 12 | 6 BL/6 LL | 8/4 | 44.8 (28–66) | 3.67 (1–5.50) | 4.68 (3.5–5.95) | Pretreatment (24-month follow-up) | |||
| de Souza et al., 2022 [22] | Brazil | Cross-sectional | II | 10 | 10 II | - | - | - | - | - |
| TT | 10 | 10 TT | - | - | - | - | - | |||
| LL | 10 | 10 LL | - | - | - | - | - | |||
| Silva et al., 2017 [5] | Brazil | Cross-sectional | T-lep | 26 | 26 BT | 14/12 | 51 (20–69) | 0 (0–0) | 0 (0–0) | 26 Pretreatment |
| L-lep | 28 | 3 BL/25 LL | 22/6 | 45.71 (21–73) | 4.33 (0.50–5.85) | 5.23 (2.70–5.90) | 28 Pretreatment | |||
| T1R | 11 | 11 BL | 7/4 | 53 (26–70) | 1.45 (0–3.75) | 2.35 (0–3.80) | 2 Pretreatment/9 on treatment | |||
| Silva et al., 2021 [23] | Brazil | Cross-sectional and cohort | PB | 14 | 14 BT | 6/8 | 54.5 (8–92) | 0 (0–0) | 0 (0–0) | - |
| MB No Progression | 8 | 4 BL/4 LL | 7/1 | 53.37 (34–65) | 2.15 (1.50–5.50) | 4.38 (2.85–5.95) | - | |||
| MB Progression | 7 | 4 BL/3 LL | 5/2 | 45.14 (32–69) | 2.98 (0.50–4.67) | 4.6 (2.7–5.95) | - | |||
| T1R | 12 | 9 BL/3 LL | 9/3 | 49.16 (17–66) | 2.66 (0.75–5.85) | 2.06 (0–3.8) | - | |||
| Yang et al., 2014a [24] | China | Cross-sectional | Healthy Control | 432 | - | 302/163 | 57.1 ± 7.2 | - | - | - |
| Leprosy Cases | 412 | 79 PB/333 MB | 291/141 | 56.8 ± 6.8 | - | - | - | |||
| Yang et al., 2014b [25] | China | Cross- sectional | Healthy Control | 46 | - | 30/16 | - | - | - | - |
| Leprosy Cases | 78 | 9 TT/25 BT/28 BL/16 LL | 52/26 | - | - | - | - |
| Author, Year [Ref.] | Sample | Characteristics |
|---|---|---|
| Barbosa et al., 2018 [21] | Skin Lesion Macrophages Isolated PBMCs and Monocyte Cultures | MB patients Healthy donors (+ armadillo g-irradiated M. leprae) |
| Ma et al., 2017 [19] | Isolated PBMCs and Monocyte Cultures | Healthy donors (6 males) + live or killed Thai53- strain M. leprae |
| Ma et al., 2018 [26] | Isolated PBMCs and Monocyte Cultures | Healthy donor (1 female) + live or killed M. leprae strain from 2 T-lep and 6 L-lep patients |
| Silva et al., 2017 [5] | Skin Lesion Macrophages Differentiated Macrophages | T-lep, L-lep, and T1R patients Human monocytic cell line THP-1 obtained from the American Type Culture Collection |
| Isolated PBMCs and Monocyte Cultures | Healthy donors | |
| Silva et al., 2021 [23] | Skin Biopsies Differentiated Macrophages | PB, MB, and T1R patients Human monocytic cell line THP-1 obtained from the American Type Culture Collection |
| Isolated PBMCs and Monocyte Cultures | Healthy donors | |
| Yang et al., 2014a [24] | Isolated PBMCs | Healthy donors + heat-killed T-58-strain M. leprae |
| Yang et al., 2014b [25] | Isolated PBMCs and CD4+ T Cells, Monocytes and Macrophages Cultures | Healthy donors + heat-killed T-58-strain M. leprae |
| Author, Year [Ref.] | Leprosy Patients | Cell Type | Autophagy Outcomes |
|---|---|---|---|
| Barbosa et al., 2018 [21] | Multibacillary with reversal reaction (24-month follow-up) | Skin lesion cells and PBMCs | ↓ LC3 mRNA and several autophagic process-related genes associated with ↑ TLR2 and MLST8. ↑ NLRP3, CASP1, and IL1B mRNA levels, and ↑ IL-1β serum concentration. |
| de Sousa et al., 2022 [22] | Indeterminate (II), tuberculoid (TT), and lepromatous (LL) | Skin lesion samples | ↑ FasL, caspase-8, RIP1 and RIP3, MLKL, BAX, caspase-3, and caspase-1 in the LL form. ↓ Beclin-1 in the LL and II forms, ↑ in the TT form. |
| Ma et al., 2017 [19] | In vitro Cell cultures + live or killed M. leprae stimuli | Monocytes and T lymphocytes from PBMCS | Killed M. leprae infection induced production of proinflammatory IL-1β, IL-6, IL-12 and TNF-α, which ↑ xenophagy. Live M. leprae infection also ↑ xenophagy, primed anti-inflammatory T cell responses by ↑ IL-10 which ↓ xenophagy. |
| Ma et al., 2018 [26] | In vitro Cell cultures + live or killed M. leprae strains from Tuberculoid (T-lep) and Lepromatous (L-lep) leprosy patients | Monocytes and T lymphocytes from PBMCs | ↑ IRGM and IL-12 expression in macrophages treated by killed M. leprae strains, which ↑ xenophagy. ↓ IRGM, MHC-II expression, and caspase-3 and caspase-9 activity in macrophages treated with both live M. leprae strains, which ↓ xenophagy and apoptosis. |
| Silva et al., 2017 [5] | Tuberculoid (T-lep) and lepromatous (L-lep) leprosy patients and type 1 reaction (T1R) patients | Skin lesion cells, human monocytic cell line THP-1, and PBMCs | ↑ LC3-II levels via immunofluorescence and BECN1, GPSM3, ATG14, APOL1 e TPR gene expression in T-lep patients. ↑ LC3-I levels in L-lep patients by immunofluorescence, and ↑ BCL2 expression which ↓ xenophagy. ↑ IFN-γ and restored xenophagy levels in L-lep patients who developed the reversal reaction. |
| Silva et al., 2021 [23] | Paucibacillary (PB), multibacillary (MB) and type 1 reaction (T1R) patients (24-month follow-up) | Skin biopsies, human monocytic cell line THP-1, and PBMCs | ↑ autophagic process-related genes: RPTOR, ULK2, ATG16L2, ATG10, ATG7, FKBP15, GPSM1, GPSM2, SEC23B, SQSTM1, and LAMP2 in M. leprae-stimulated THP-1 cells in the presence of IFN-γ, and ↑ IL-15 secretion. ↑ IL15 mRNA levels in T1R lesions compared to PB and MB groups. Presence of 13 common autophagic genes (FRS3, GFI1B, GNAI3, GPSM1, GPSM2, LETM2, RASD1, RPTOR, SEC23B, SEC24A, TPR, UVRAG, and BECN2) between T1R skin biopsies and stimulated THP-1cells with M. leprae and IFN-γ. |
| Yang et al., 2014a [24] | Paucibacillary and multibacillary | PBMCs | IRGM polymorphism (rs13361189TC and CC genotypes) is associated with ↑ susceptibility to leprosy, and rs13361189CC genotype ↑ leprosy complications. ↑ IFN-γ and IL-4 in M. leprae-infected PBMC with rs13361189CC genotype. No differences in the distribution of rs13361189 SNP between paucibacillary and multibacillary forms. |
| Yang et al., 2014b [25] | Lepromatous lepromatous (LL), borderline lepromatous (BL), borderline tuberculoid (BT), and tuberculoid (TT) | CD4+ T cells, monocytes, and monocyte-derived macrophages from PBMCs | ↑ IRGM protein and mRNA levels in monocytes and macrophages upon stimulation with M. leprae. ↑ IRGM in monocytes from TT type, then BT type, BL type, and followed by LL type, showing inverse correlation with the severity of the disease. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerqueira, D.D.N.; Pereira, A.L.S.; da Costa, A.E.C.; de Souza, T.J.; de Sousa Fernandes, M.S.; Souto, F.O.; Santos, P.d.A. Xenophagy as a Strategy for Mycobacterium leprae Elimination during Type 1 or Type 2 Leprosy Reactions: A Systematic Review. Pathogens 2023, 12, 1455. https://doi.org/10.3390/pathogens12121455
Cerqueira DDN, Pereira ALS, da Costa AEC, de Souza TJ, de Sousa Fernandes MS, Souto FO, Santos PdA. Xenophagy as a Strategy for Mycobacterium leprae Elimination during Type 1 or Type 2 Leprosy Reactions: A Systematic Review. Pathogens. 2023; 12(12):1455. https://doi.org/10.3390/pathogens12121455
Chicago/Turabian StyleCerqueira, Débora Dantas Nucci, Ana Letícia Silva Pereira, Ana Elisa Coelho da Costa, Tarcísio Joaquim de Souza, Matheus Santos de Sousa Fernandes, Fabrício Oliveira Souto, and Patrícia d’Emery Alves Santos. 2023. "Xenophagy as a Strategy for Mycobacterium leprae Elimination during Type 1 or Type 2 Leprosy Reactions: A Systematic Review" Pathogens 12, no. 12: 1455. https://doi.org/10.3390/pathogens12121455







