Integrating Transcriptomics and Metabolomics to Explore the Novel Pathway of Fusobacterium nucleatum Invading Colon Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Fusobacterium nucleatum ATCC25586
2.2. Isolation of Fnevs
2.3. Transmission Electron Microscopy (TEM)
2.4. Scanning Electron Microscopy (SEM)
2.5. Nanoparticle Tracking Analysis
2.6. Protein Identification and Analysis of Fnevs
2.7. Cell Source and Culture
2.8. Fluorescent Labeling Fnevs
2.9. Detection of Cell Proliferation by CCK-8
2.10. Flow Cytometry Analysis of Apoptosis
2.11. Detection of Intracellular ROS Level and SOD Enzyme Activity
2.12. Wound-Healing Assay and Transwell Assay for Cell Migration and Invasion
2.13. Transcriptome Sequencing (RNA-Seq) and Analysis
2.14. LC-MS/MS for Differential Metabolite Analysis
2.15. Statistical Analysis
3. Results
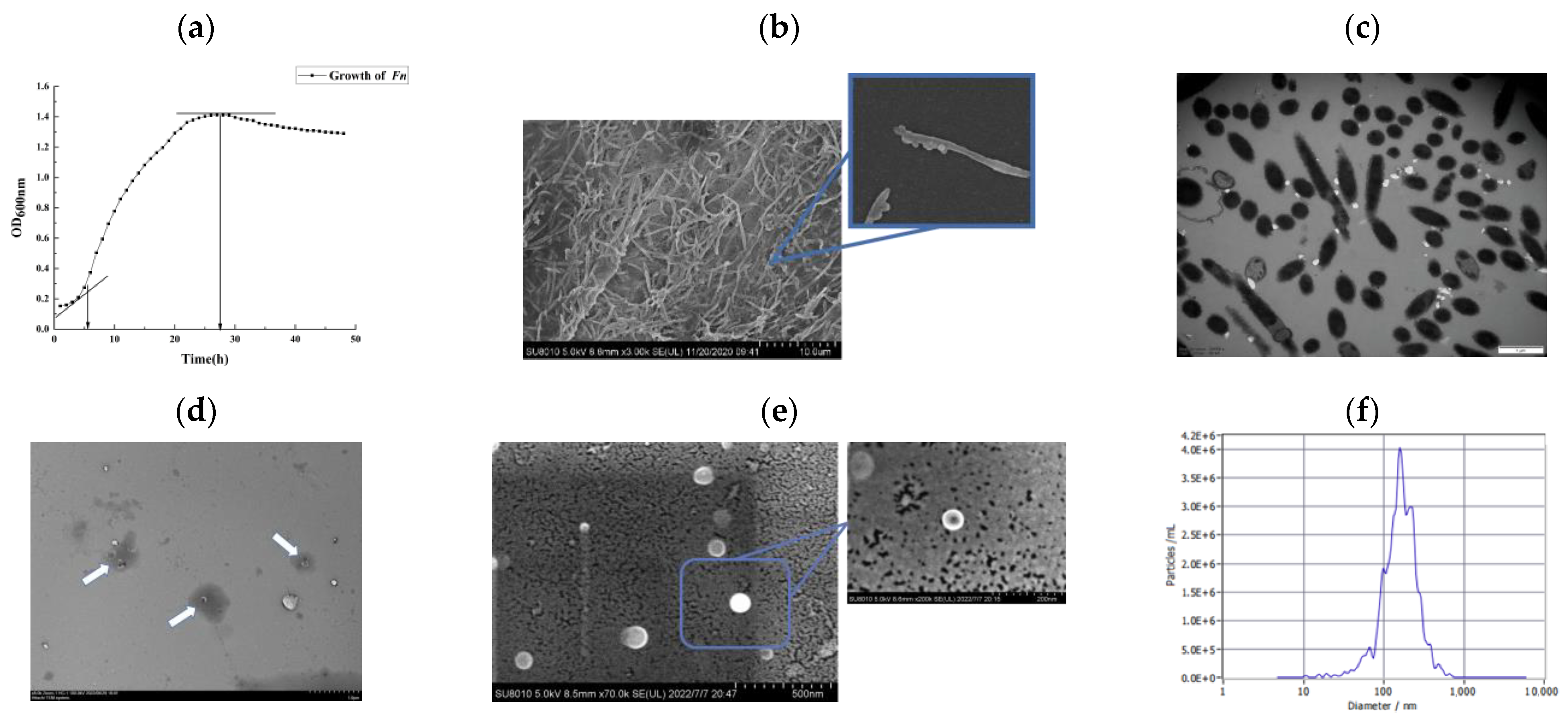
3.1. Growth Kinetic Curve and Structural Observation of Fusobacterium nucleatum ATCC25586
3.2. Characterization of Extracellular Vesicles of Fusobacterium nucleatum ATCC25586 (Fnevs)
3.3. Protein Identification of Fnevs
3.4. Internalization of Fnevs
3.5. Effect of Fnevs on the Proliferation and Apoptosis of Colon Cancer Cells
3.6. Effect of Fnevs on the Migration, Invasion, and Healing Ability of Colon Cancer Cells
3.7. Effect of Fnevs on the ROS Level and SOD Enzyme Activity in Colon Cancer Cells
3.8. Differential Gene Expression and Enrichment Pathways in Fnevs-Treated Colon Cancer Cells
3.9. Differential Metabolites and Metabolic Pathways in Fnevs-Treated Colon Cancer Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; A Hermes, G.D.; Hirschfield, G.M.; Hold, G.L.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.Q.; Jha, R.; Li, A.O.; Liu, H.W.; Zhang, Z.; Zhang, C.C.; Zhai, Q.X.; Zhang, J.C. Probiotics (Lactobacillus Plantarum Hnu082) Supplementation Relieves Ulcerative Colitis by Affecting Intestinal Barrier Functions, Immunity-Related Gene Expression, Gut Microbiota, and Metabolic Pathways in Mice. Microbiol. Spectr. 2022, 10, e01651-22. [Google Scholar] [CrossRef] [PubMed]
- Toscano, M.; De Grandi, R.; Stronati, L.; De Vecchi, E.; Drago, L. Effect of Lactobacillus rhamnosus HN001 and Bifidobacterium longum BB536 on the healthy gut microbiota composition at phyla and species level: A preliminary study. World J. Gastroenterol. 2017, 23, 2696–2704. [Google Scholar] [CrossRef] [PubMed]
- Kosumi, K.; Hamada, T.; Koh, H.; Borowsky, J.; Bullman, S.; Twombly, T.S.; Nevo, D.; Masugi, Y.; Liu, L.; da Silva, A.; et al. The Amount of Bifidobacterium Genus in Colorectal Carcinoma Tissue in Relation to Tumor Characteristics and Clinical Outcome. Am. J. Pathol. 2018, 188, 2839–2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolowschiak, T.; Mueller, A.A.; Pisan, L.J.; Feigelman, R.; Felmy, B.; Sellin, M.E.; Namineni, S.; Nguyen, B.D.; Wotzka, S.Y.; Heikenwalder, M.; et al. Ifn-Gamma Hinders Recovery from Mucosal Inflammation During Antibiotic Therapy for Salmonella Gut Infection. Cell Host Microbe 2016, 20, 238–249. [Google Scholar] [CrossRef] [Green Version]
- Gagnaire, A.; Nadel, B.; Raoult, D.; Neefjes, J.; Gorvel, J.-P. Collateral damage: Insights into bacterial mechanisms that predispose host cells to cancer. Nat. Rev. Genet. 2017, 15, 109–128. [Google Scholar] [CrossRef]
- Ruano-Gallego, D.; Sanchez-Garrido, J.; Kozik, Z.; Núñez-Berrueco, E.; Cepeda-Molero, M.; Mullineaux-Sanders, C.; Clark, J.N.-B.; Slater, S.L.; Wagner, N.; Glegola-Madejska, I.; et al. Type III secretion system effectors form robust and flexible intracellular virulence networks. Science 2021, 371, eabc9531. [Google Scholar] [CrossRef]
- Hendrix, A.; De Wever, O. Systemically Circulating Bacterial Extracellular Vesicles: Origin, Fate, and Function. Trends Microbiol. 2022, 30, 213–316. [Google Scholar] [CrossRef]
- Tsatsaronis, J.A.; Franch-Arroyo, S.; Resch, U.; Charpentier, E. Extracellular Vesicle Rna: A Universal Mediator of Microbial Communication? Trends Microbiol. 2018, 26, 401–410. [Google Scholar] [CrossRef]
- Díaz-Garrido, N.; Badia, J.; Baldomà, L. Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles 2021, 10, e12161. [Google Scholar] [CrossRef] [PubMed]
- Morishita, M.; Horita, M.; Higuchi, A.; Marui, M.; Katsumi, H.; Yamamoto, A. Characterizing Different Probiotic-Derived Extracellular Vesicles as a Novel Adjuvant for Immunotherapy. Mol. Pharm. 2021, 18, 1080–1092. [Google Scholar] [CrossRef]
- Raftar, S.K.A.; Ashrafian, F.; Yadegar, A.; Lari, A.; Moradi, H.R.; Shahriary, A.; Azimirad, M.; Alavifard, H.; Mohsenifar, Z.; Davari, M.; et al. The Protective Effects of Live and Pasteurized Akkermansia muciniphila and Its Extracellular Vesicles against HFD/CCl4-Induced Liver Injury. Microbiol. Spectr. 2021, 9, e00484-21. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Choi, Y.; Kim, D.-K.; Park, H.T.; Ghim, J.; Kwon, Y.; Jeon, J.; Kim, M.-S.; Jee, Y.-K.; Gho, Y.S.; et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp. Mol. Med. 2018, 50, e450. [Google Scholar] [CrossRef] [Green Version]
- Tong, L.; Zhang, X.; Hao, H.; Liu, Q.; Zhou, Z.; Liang, X.; Liu, T.; Gong, P.; Zhang, L.; Zhai, Z.; et al. Lactobacillus rhamnosus GG Derived Extracellular Vesicles Modulate Gut Microbiota and Attenuate Inflammatory in DSS-Induced Colitis Mice. Nutrients 2021, 13, 3319. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Zhang, X.; Tong, L.; Liu, Q.; Liang, X.; Bu, Y.; Gong, P.; Liu, T.; Zhang, L.; Xia, Y.; et al. Effect of Extracellular Vesicles Derived From Lactobacillus plantarum Q7 on Gut Microbiota and Ulcerative Colitis in Mice. Front. Immunol. 2021, 12, 777147. [Google Scholar] [CrossRef]
- Lee, J.; Kim, O.Y.; Gho, Y.S. Proteomic Profiling of Gram-Negative Bacterial Outer Membrane Vesicles: Current Perspectives. Proteom. Clin. Appl. 2016, 10, 897–909. [Google Scholar] [CrossRef]
- Choi, D.-S.; Kim, D.-K.; Choi, S.J.; Lee, J.; Choi, J.-P.; Rho, S.; Park, S.-H.; Kim, Y.-K.; Hwang, D.; Gho, Y.S. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics 2011, 11, 3424–3429. [Google Scholar] [CrossRef]
- Shangchun, G.; Zhao, L.; Tao, S.; Zhang, C. Research Progress on the Role of Extracellular Vesicles in Bacterial Pathogenesis. Chin. J. Reparative Reconstr. Surg. 2018, 32, 1597–1604. [Google Scholar]
- Brennan, C.A.; Garrett, W.S. Fusobacterium Nucleatum–Symbiont, Opportunist and Oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Parhi, L.; Abed, J.; Shhadeh, A.; Alon-Maimon, T.; Udi, S.; Ben-Arye, S.L.; Tam, J.; Parnas, O.; Padler-Karavani, V.; Goldman-Wohl, D.; et al. Placental Colonization by Fusobacterium Nucleatum Is Mediated by Binding of the Fap2 Lectin to Placentally Displayed Gal-Galnac. Cell Rep. 2022, 38, 110537. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Fang, J.-Y. Fusobacterium nucleatum, a key pathogenic factor and microbial biomarker for colorectal cancer. Trends Microbiol. 2022, 31, 159–172. [Google Scholar] [CrossRef]
- Mohammadi, M.; Mirzaei, H.; Motallebi, M. The role of anaerobic bacteria in the development and prevention of colorectal cancer: A review study. Anaerobe 2022, 73, 102501. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Z.; Weng, W.H.; Peng, J.J.; Hong, L.M.; Yang, L.; Toiyama, Y.; Gao, R.Y.; Liu, M.F.; Yin, M.M.; Pan, C.; et al. Fusobacterium Nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-Kappa B, and up-Regulating Expression of Microrna-21. Gastroenterology 2017, 152, 851. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, L.; Zheng, S.; Li, M.J.; Xu, C.C.; Jia, D.J.C.; Qi, Y.D.; Hou, T.Y.; Wang, L.; Wang, B.Y.; et al. Fusobacterium Nucleatum Promotes Colorectal Cancer Cells Adhesion to Endothelial Cells and Facilitates Extravasation and Metastasis by Inducing Alpk1/Nf-Kappa B/Icam1 Axis. Gut Microbes 2022, 14, 2038852. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Chen, Y.; Cao, P.; Su, W.H.; Zhan, N.; Dong, W.G. Fusobacterium Nucleatum Facilitates Ulcerative Colitis through Activating Il-17f Signaling to Nf-Kappa B Via the Upregulation of Card3 Expression. J. Pathol. 2020, 250, 170–182. [Google Scholar] [CrossRef]
- Meng, Q.; Gao, Q.; Mehrazarin, S.; Tangwanichgapong, K.; Wang, Y.; Huang, Y.; Pan, Y.; Robinson, S.; Liu, Z.; Zangiabadi, A.; et al. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 2021, 22, e52891. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Baik, J.E.; Lagana, S.M.; Han, R.P.; Raab, W.J.; Sahoo, D.; Dalerba, P.; Wang, T.C.; Han, Y.W. Fusobacterium Nucleatum Promotes Colorectal Cancer by Inducing Wnt/Beta-Catenin Modulator Annexin A1. EMBO Rep. 2019, 20, e47638. [Google Scholar] [CrossRef] [PubMed]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Rockman, S.P.; Currie, S.A.; Ciavarella, M.; Vincan, E.; Dow, C.; Thomas, R.J.; Phillips, W.A. Id2 Is a Target of the Beta-Catenin/T Cell Factor Pathway in Colon Carcinoma. J. Biol. Chem. 2001, 276, 45113–45119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talaat, I.M.; Yakout, N.M.; Soliman, A.S.; Venkatachalam, T.; Vinod, A.; Eldohaji, L.; Nair, V.; Hareedy, A.; Kandil, A.; Abdel-Rahman, W.M.; et al. Evaluation of Galanin Expression in Colorectal Cancer: An Immunohistochemical and Transcriptomic Study. Front. Oncol. 2022, 12, 877147. [Google Scholar] [CrossRef] [PubMed]
- Goyal, A.; Duke, E.R.; Cardozo-Ojeda, E.F.; Schiffer, J.T. Modeling Explains Prolonged Sars-Cov-2 Nasal Shedding Relative to Lung Shedding in Remdesivir-Treated Rhesus Macaques. Iscience 2022, 25, 104448. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Yang, M.; Li, Y.; Yang, F.; Feng, Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Transfer Wnt4 to Normoxic Cells to Elicit a Prometastatic Phenotype. Int. J. Biol. Sci. 2018, 14, 2094–2102. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Zeng, Q.H.; Cao, P.G.; Xie, D.; Chen, X.; Yang, F.; He, L.Y.; Dai, Y.B.; Li, J.J.; Liu, X.M.; et al. Ripk4 Promotes Bladder Urothelial Carcinoma Cell Aggressiveness by Upregulating Vegf-a through the Nf-Kappa B Pathway. Br. J. Cancer 2018, 118, 1617–1627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclerc, D.; Pham, D.N.T.; Lévesque, N.; Truongcao, M.; Foulkes, W.D.; Sapienza, C.; Rozen, R. Oncogenic role of PDK4 in human colon cancer cells. Br. J. Cancer 2017, 116, 930–936. [Google Scholar] [CrossRef]
- Huynh, J.; Baloyan, D.; Chisanga, D.; Shi, W.; O’Brien, M.; Afshar-Sterle, S.; Alorro, M.; Pang, L.; Williams, D.S.; Parslow, A.C.; et al. Host IL11 Signaling Suppresses CD4+ T cell–Mediated Antitumor Responses to Colon Cancer in Mice. Cancer Immunol. Res. 2021, 9, 735–747. [Google Scholar] [CrossRef]
- Wang, W.; Shen, T.; Dong, B.; Creighton, C.J.; Meng, Y.; Zhou, W.; Shi, Q.; Zhou, H.; Zhang, Y.; Moore, D.D.; et al. MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J. Clin. Investig. 2019, 129, 1015–1029. [Google Scholar] [CrossRef]
- Middel, P.; Reich, K.; Polzien, F.; Blaschke, V.; Hemmerlein, B.; Herms, J.; Korabiowska, M.; Radzun, H.J. Interleukin 16 Expression and Phenotype of Interleukin 16 Producing Cells in Crohn’s Disease. GUT 2001, 49, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-K.; Zhao, H.-M.; Wang, H.-Y.; Ge, W.; Zhong, Y.-B.; Long, J.; Liu, D.-Y. Regulatory Effect of Sishen Pill on Tfh Cells in Mice With Experimental Colitis. Front. Physiol. 2020, 11, 589. [Google Scholar] [CrossRef]
- Kakiuchi, N.; Yoshida, K.; Uchino, M.; Kihara, T.; Akaki, K.; Inoue, Y.; Kawada, K.; Nagayama, S.; Yokoyama, A.; Yamamoto, S.; et al. Frequent mutations that converge on the NFKBIZ pathway in ulcerative colitis. Nature 2020, 577, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.-P.; Jiao, H.-L.; Wang, S.-Y.; Xiao, Z.-Y.; Zhang, D.; Qiu, J.-F.; Zhang, L.-J.; Zhao, Y.-L.; Li, T.-T.; Liang, L.; et al. Hypermethylation of DMTN promotes the metastasis of colorectal cancer cells by regulating the actin cytoskeleton through Rac1 signaling activation. J. Exp. Clin. Cancer Res. 2018, 37, 299. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-J.; Lee, J.; Li, K.K.; Holland, D.; Maughan, H.; Guttman, D.S.; Yusta, B.; Drucker, D.J. Disruption of the Murine Glp2r Impairs Paneth Cell Function and Increases Susceptibility to Small Bowel Enteritis. Endocrinology 2012, 153, 1141–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oertel, S.; Scholich, K.; Weigert, A.; Thomas, D.; Schmetzer, J.; Trautmann, S.; Wegner, M.-S.; Radeke, H.H.; Filmann, N.; Brüne, B.; et al. Ceramide synthase 2 deficiency aggravates AOM-DSS-induced colitis in mice: Role of colon barrier integrity. Cell. Mol. Life Sci. 2017, 74, 3039–3055. [Google Scholar] [CrossRef]
- Lautaoja, J.H.; Lalowski, M.; Nissinen, T.A.; Hentilä, J.J.; Shi, Y.; Ritvos, O.; Cheng, S.; Hulmi, J.J. Muscle and serum metabolomes are dysregulated in colon-26 tumor-bearing mice despite amelioration of cachexia with activin receptor type 2B ligand blockade. Am. J. Physiol. Metab. 2019, 316, E852–E865. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Schwörer, S.; Berisa, M.; Kyung, Y.J.; Ryu, K.W.; Yi, J.; Jiang, X.; Cross, J.R.; Thompson, C.B. Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science 2021, 372, 968–972. [Google Scholar] [CrossRef]
- Sesardic, D.; Boobis, A.; Murray, B.; Murray, S.; Segura, J.; de la Torre, R.; Davies, D. Furafylline is a potent and selective inhibitor of cytochrome P450IA2 in man. Br. J. Clin. Pharmacol. 1990, 29, 651–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef]
- Garcia-Barros, M.; Coant, N.; Truman, J.P.; Snider, A.J.; Hannun, Y.A. Sphingolipids in Colon Cancer. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2014, 1841, 773–782. [Google Scholar] [CrossRef] [Green Version]
- Spector, T. Oxypurinol as an inhibitor of xanthine oxidase-catalyzed production of superoxide radical. Biochem. Pharmacol. 1988, 37, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Pudakalakatti, S.; Titus, M.; Enriquez, J.S.; Ramachandran, S.; Zacharias, N.M.; Shureiqi, I.; Liu, Y.; Yao, J.C.; Zuo, X.; Bhattacharya, P.K. Identifying the Metabolic Signatures of PPARD-Overexpressing Gastric Tumors. Int. J. Mol. Sci. 2022, 23, 1645. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, R.; Zhao, L.; Lv, L.; Zhou, F.; Zhang, T.; Lu, F.; Yan, H.; Duan, G. Celastrol Suppresses Tryptophan Catabolism in Human Colon Cancer Cells as Revealed by Metabolic Profiling and Targeted Metabolite Analysis. Biol. Pharm. Bull. 2018, 41, 1243–1250. [Google Scholar] [CrossRef] [Green Version]
- Matsui, T.; Ichikawa, H.; Fujita, T.; Takemura, S.; Takagi, T.; Osada-Oka, M.; Minamiyama, Y. Histidine and Arginine Modulate Intestinal Cell Restitution Via Transforming Growth Factor-Beta(1). Eur. J. Pharmacol. 2019, 850, 35–42. [Google Scholar] [CrossRef]
- Thangaraju, M.; Carswell, K.N.; Prasad, P.D.; Ganapathy, V. Colon Cancer Cells Maintain Low Levels of Pyruvate to Avoid Cell Death Caused by Inhibition of Hdac1/Hdac3. Biochem. J. 2009, 417, 379–389. [Google Scholar] [CrossRef]
- Wang, M.; Xia, W.; Li, H.; Liu, F.; Li, Y.Y.; Sun, X.J.; Lu, S.F.; Xu, S.Q. Normal Pregnancy Induced Glucose Metabolic Stress in a Longitudinal Cohort of Healthy Women Novel Insights Generated from a Urine Metabolomics Study. Medicine 2018, 97, e12417. [Google Scholar] [CrossRef]
- Northrop-Albrecht, E.J.; Taylor, W.R.; Huang, B.Q.; Kisiel, J.B.; Lucien, F. Assessment of extracellular vesicle isolation methods from human stool supernatant. J. Extracell. Vesicles 2022, 11, e12208. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.H.; Chen, J.; Chen, F.F.; Zeng, Q.Y.; Liu, W.L.; Zhang, G. Exosomes Derived from Fusobacterium Nucleatum-Infected Colorectal Cancer Cells Facilitate Tumour Metastasis by Selectively Carrying Mir-1246/92b-3p/27a-3p and Cxcl16. Gut 2021, 70, 1507–1519. [Google Scholar] [CrossRef]
- Bomberger, J.M.; MacEachran, D.P.; Coutermarsh, B.A.; Ye, S.; O’Toole, G.A.; Stanton, B.A. Long-Distance Delivery of Bacterial Virulence Factors by Pseudomonas aeruginosa Outer Membrane Vesicles. PLOS Pathog. 2009, 5, e1000382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chutkan, H.; Kuehn, M.J. Context-Dependent Activation Kinetics Elicited by Soluble versus Outer Membrane Vesicle-Associated Heat-Labile Enterotoxin. Infect. Immun. 2011, 79, 3760–3769. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Xu, J.; Yang, X.; Wang, D.; Xu, X. Integrating Transcriptomics and Metabolomics to Explore the Novel Pathway of Fusobacterium nucleatum Invading Colon Cancer Cells. Pathogens 2023, 12, 201. https://doi.org/10.3390/pathogens12020201
Wu X, Xu J, Yang X, Wang D, Xu X. Integrating Transcriptomics and Metabolomics to Explore the Novel Pathway of Fusobacterium nucleatum Invading Colon Cancer Cells. Pathogens. 2023; 12(2):201. https://doi.org/10.3390/pathogens12020201
Chicago/Turabian StyleWu, Xinyu, Jinzhao Xu, Xiaoying Yang, Danping Wang, and Xiaoxi Xu. 2023. "Integrating Transcriptomics and Metabolomics to Explore the Novel Pathway of Fusobacterium nucleatum Invading Colon Cancer Cells" Pathogens 12, no. 2: 201. https://doi.org/10.3390/pathogens12020201




