Pathological Features and Genomic Characterization of an Actinobacillus equuli subsp. equuli Bearing Unique Virulence-Associated Genes from an Adult Horse with Pleuropneumonia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Postmortem Examination and Sample Collection
2.2. Histopathology
2.3. Bacteriology, MALDI-TOF Mass Spectrometry, and Antibiotic Sensitivity
2.4. DNA Extraction, Whole Genome Sequencing, and Assembly
2.5. A. equuli Subsp. equuli Genome Annotation and Analysis
2.6. Phylogenetic Analysis
3. Results
3.1. Case Presentation
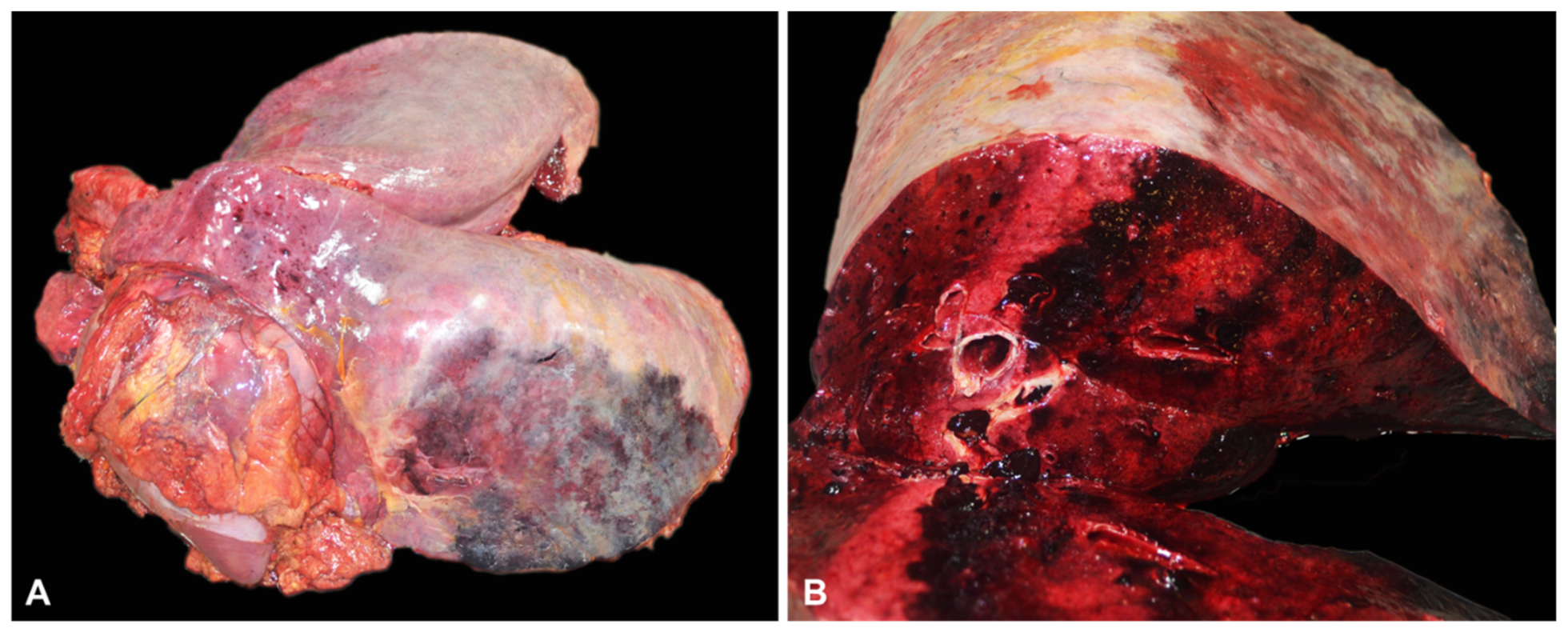
3.2. Gross and Histopathological Findings
3.3. Bacteriology, MALDI-TOF Mass Spectrometry, and Antibiotic Sensitivity
3.4. Genome Properties of the A. equuli subsp. equuli 4524 Isolate
3.5. Comparative Genome Analysis of A. equuli Subsp. equuli 4524 with ATCC 19392 Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacInnes, J.I. Actinobacillus; Wiley-Blackwell: Ames, IA, USA, 2010. [Google Scholar]
- Bujold, A.R.; Shure, A.E.; Liu, R.; Kropinski, A.M.; MacInnes, J.I. Investigation of putative invasion determinants of Actinobacillus species using comparative genomics. Genomics 2019, 111, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Kuhnert, P.; Berthoud, H.; Christensen, H.; Bisgaard, M.; Frey, J. Phylogenetic relationship of equine Actinobacillus species and distribution of RTX toxin genes among clusters. Vet. Res. 2003, 34, 353–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthews, S.; Dart, A.J.; Dowling, B.A.; Hodgson, J.L.; Hodgson, D.R. Peritonitis associated with Actinobacillus equuli in horses: 51 cases. Aust. Vet. J. 2001, 79, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Aalbaek, B.; Ostergaard, S.; Buhl, R.; Jensen, H.E.; Christensen, H.; Bisgaard, M. Actinobacillus equuli subsp. equuli associated with equine valvular endocarditis. APMIS 2007, 115, 1437–1442. [Google Scholar] [CrossRef]
- Castagnetti, C.; Rossi, M.; Parmeggiani, F.; Zanoni, R.G.; Pirrone, A.; Mariella, J. Facial cellulitis due to Actinobacillus equuli infection in a neonatal foal. Vet. Rec. 2008, 162, 347–349. [Google Scholar] [CrossRef]
- Donahue, J.M.; Sells, S.F.; Bolin, D.C. Classification of Actinobacillus spp isolates from horses involved in mare reproductive loss syndrome. Am. J. Vet. Res. 2006, 67, 1426–1432. [Google Scholar] [CrossRef]
- Stewart, A.J.; Hinchcliff, K.W.; Saville, W.J.; Jose-Cunilleras, E.; Hardy, J.; Kohn, C.W.; Reed, S.M.; Kowalski, J.J. Actinobacillus sp. bacteremia in foals: Clinical signs and prognosis. J. Vet. Intern. Med. 2002, 16, 464–471. [Google Scholar] [CrossRef]
- Ward, C.L.; Wood, J.L.; Houghton, S.B.; Mumford, J.A.; Chanter, N. Actinobacillus and Pasteurella species isolated from horses with lower airway disease. Vet. Rec. 1998, 143, 277–279. [Google Scholar] [CrossRef]
- Edwards, P.R. Studies on Shigella equirulis (Bact. viscosum equi). Bulletin- Kentucky, Agricultural Experiment Station, 1931; 289–330.11. [Google Scholar]
- Sternberg, S.; Brandstrom, B. Biochemical fingerprinting and ribotyping of isolates of Actinobacillus equuli from healthy and diseased horses. Vet. Microbiol. 1999, 66, 53–65. [Google Scholar] [CrossRef]
- Layman, Q.D.; Rezabek, G.B.; Ramachandran, A.; Love, B.C.; Confer, A.W. A retrospective study of equine actinobacillosis cases: 1999–2011. J. Vet. Diagn. Investig. 2014, 26, 365–375. [Google Scholar] [CrossRef]
- Benavente, C.E.; Fuentealba, I.C. Actinobacillus suis and Actinobacillus equuli, emergent pathogens of septic embolic nephritis, a new challenge for the swine industry. Arch. Med. Vet. 2012, 44, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Thompson, A.B.; Postey, R.C.; Snider, T.; Pasma, T. Actinobacillus equuli as a primary pathogen in breeding sows and piglets. Can. Vet. J. 2010, 51, 1223–1225. [Google Scholar]
- Pusterla, N.; Jones, M.E.; Mohr, F.C.; Higgins, J.K.; Mapes, S.; Jang, S.S.; Samitz, E.M.; Byrne, B.A. Fatal pulmonary hemorrhage associated with RTX toxin producing Actinobacillus equuli subspecies haemolyticus infection in an adult horse. J. Vet. Diagn. Investig. 2008, 20, 118–121. [Google Scholar] [CrossRef] [Green Version]
- Bosse, J.T.; Janson, H.; Sheehan, B.J.; Beddek, A.J.; Rycroft, A.N.; Kroll, J.S.; Langford, P.R. Actinobacillus pleuropneumoniae: Pathobiology and pathogenesis of infection. Microbes Infect. 2002, 4, 225–235. [Google Scholar] [CrossRef]
- Bujold, A.R.; MacInnes, J.I. Identification of putative adhesins of Actinobacillus suis and their homologues in other members of the family Pasteurellaceae. BMC Res. Notes 2015, 8, 675. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genom. Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Resm. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Blom, J.; Albaum, S.P.; Doppmeier, D.; Puhler, A.; Vorholter, F.J.; Zakrzewski, M.; Goesmann, A. EDGAR: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinform. 2009, 10, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Richter, M.; Rossello-Mora, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Goker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Schmid, J.; Heider, D.; Wendel, N.J.; Sperl, N.; Sieber, V. Bacterial Glycosyltransferases: Challenges and Opportunities of a Highly Diverse Enzyme Class Toward Tailoring Natural Products. Front. Microbiol. 2016, 7, 182. [Google Scholar] [CrossRef] [Green Version]
- Berthoud, H.; Frey, J.; Kuhnert, P. Characterization of Aqx and its operon: The hemolytic RTX determinant of Actinobacillus equuli. Vet. Microbiol. 2002, 87, 159–174. [Google Scholar] [CrossRef]
- Kuhnert, P.; Berthoud, H.; Straub, R.; Frey, J. Host cell specific activity of RTX toxins from haemolytic Actinobacillus equuli and Actinobacillus suis. Vet. Microbiol. 2003, 92, 161–167. [Google Scholar] [CrossRef]
- Frey, J. The role of RTX toxins in host specificity of animal pathogenic Pasteurellaceae. Vet. Microbiol. 2011, 153, 51–58. [Google Scholar] [CrossRef]
- Bannerman, D.D.; Goldblum, S.E. Mechanisms of bacterial lipopolysaccharide-induced endothelial apoptosis. Am. J. Physiol. Lung Cell Mol. Physiol. 2003, 284, L899–L914. [Google Scholar] [CrossRef] [Green Version]
- Paradis, S.E.; Dubreuil, D.; Rioux, S.; Gottschalk, M.; Jacques, M. High-molecular-mass lipopolysaccharides are involved in Actinobacillus pleuropneumoniae adherence to porcine respiratory tract cells. Infect. Immun. 1994, 62, 3311–3319. [Google Scholar] [CrossRef] [Green Version]
- Jacques, M.; Paradis, S.E. Adhesin-receptor interactions in Pasteurellaceae. FEMS Microbiol. Rev. 1998, 22, 45–59. [Google Scholar] [CrossRef]
- Beddek, A.J.; Sheehan, B.J.; Bosse, J.T.; Rycroft, A.N.; Kroll, J.S.; Langford, P.R. Two TonB systems in Actinobacillus pleuropneumoniae: Their roles in iron acquisition and virulence. Infect. Immun. 2004, 72, 701–708. [Google Scholar] [CrossRef] [Green Version]
- Postle, K. TonB protein and energy transduction between membranes. J. Bioenerg. Biomembr. 1993, 25, 591–601. [Google Scholar] [CrossRef]
- Jacques, M. Surface polysaccharides and iron-uptake systems of Actinobacillus pleuropneumoniae. Can. J. Vet. Res. 2004, 68, 81–85. [Google Scholar]




| Antimicrobic | Result | Interpretation |
|---|---|---|
| Amikacin | =16 | Susceptible |
| Ampicillin | ≤0.25 | No Interpretation |
| Azithromycin | ≤0.25 | No Interpretation |
| Cefazolin | ≤4 | No Interpretation |
| Ceftazidime | ≤1 | Susceptible |
| Ceftiofur | ≤0.25 | No Interpretation |
| Chloramphenicol | ≤4 | Susceptible |
| Clarithromycin | ≤1 | No Interpretation |
| Doxycycline | ≤2 | Susceptible |
| Enrofloxacin | ≤0.25 | No Interpretation |
| Erythromycin | =0.5 | No Interpretation |
| Gentamicin | =8 | Intermediate |
| Imipenem | ≤1 | Susceptible |
| Oxacillin | ≤0.25 | No Interpretation |
| Penicillin | =0.25 | No Interpretation |
| Rifampicin | =4 | No Interpretation |
| Tetracycline | ≤2 | No Interpretation |
| Ticarcillin | ≤8 | No Interpretation |
| Ticarcillin-Clavulanate | ≤8 | Susceptible |
| Trimethoprim-sulfamethoxazole | ≤0.5 | Susceptible |
| Attribute | Value |
|---|---|
| Genome size (bp) | 2,464,726 |
| Total genes | 2362 |
| Protein coding genes | 2277 |
| RNA genes | 85 |
| tRNA genes | 61 |
| tmRNA genes | 1 |
| rRNA genes | 8 |
| ncRNA genes | 15 |
| Genes assigned to COGs | 1419 |
| CRISPR repeats | 2 |
| Code | Value | Description |
|---|---|---|
| J | 203 | Translation, ribosomal structure, and biogenesis |
| A | 1 | RNA processing and modification |
| K | 83 | Transcription |
| L | 89 | Replication, recombination, and repair |
| B | 0 | Chromatin structure and dynamics |
| D | 33 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | Nuclear structure |
| V | 30 | Defense mechanisms |
| T | 36 | Signal transduction mechanisms |
| M | 148 | Cell wall/membrane/envelope biogenesis |
| N | 6 | Cell motility |
| Z | 0 | Cytoskeleton |
| W | 1 | Extracellular structures |
| U | 28 | Intracellular trafficking, secretion, and vesicular transport |
| O | 88 | Posttranslational modification, protein turnover, chaperones |
| C | 104 | Energy production and conversion |
| G | 127 | Carbohydrate transport and metabolism |
| E | 168 | Amino acid transport and metabolism |
| F | 68 | Nucleotide transport and metabolism |
| H | 100 | Coenzyme transport and metabolism |
| I | 42 | Lipid transport and metabolism |
| P | 108 | Inorganic ion transport and metabolism |
| Q | 8 | Secondary metabolites biosynthesis, transport, and catabolism |
| R | 69 | General function prediction only |
| X | 4 | Mobilome: prophages, transposons |
| S | 78 | Function unknown |
| Species | Strain | ANI (%) | GGDC | DDH | Prob. DDH ≥ 70% |
|---|---|---|---|---|---|
| A. capsulatus | DSM 19761 | 92.7 | 0.0739 | 49 | 16.6 |
| A. delphinicola | NCTC12871 | 70.5 | 0.1742 | 25 | 0.01 |
| A. equuli subsp. equuli | ATCC 19392 | 95.9 | 0.0418 | 66.1 | 70 |
| A. indolicus | 46K2C | 74.5 | 0.2126 | 20.7 | 0 |
| A. lignieresii | NCTC4189 | 86.8 | 0.1345 | 31.5 | 0.19 |
| A. minor | NM305 | 76.4 | 0.1936 | 22.6 | 0 |
| A. pleuropneumoniae | NCTC10976 | 86.5 | 0.1335 | 31.7 | 0.2 |
| A. porcinus | NM319 | 73 | 0.1785 | 24.4 | 0.01 |
| A. porcitonsillarum | 9953L55 | 76.3 | 0.193 | 22.7 | 0 |
| A. seminis | NCTC10851 | 71.8 | 0.1857 | 23.5 | 0 |
| A. succinogenes | GXAS137 | 71.8 | 0.1847 | 23.7 | 0 |
| A. suis | ATCC33415 | 93.2 | 0.0717 | 50 | 19 |
| A. ureae | NCTC10220 | 92.7 | 0.0728 | 49.5 | 17.8 |
| Virulence Factor | Number of Shared Genes in 4524 and ATCC 19392 | Number of Genes Specific to ATCC 19392 | Number of Genes Specific to 4524 |
|---|---|---|---|
| Iron acquisition | 40 | 1 | 3 |
| LPS | 41 | 3 | 2 |
| CPS | 18 | 1 | 1 |
| Miscellaneous | 15 | 2 | 1 |
| OMP | 4 | 0 | 0 |
| Salicylic acid | 2 | 0 | 0 |
| RTX toxin | 1 | 0 | 0 |
| Autotransporter | 1 | 0 | 0 |
| Total | 122 | 7 | 7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamali, M.; Carossino, M.; Del Piero, F.; Peak, L.; Mitchell, M.S.; Willette, J.; Baker, R.; Li, F.; Kenéz, Á.; Balasuriya, U.B.R.; et al. Pathological Features and Genomic Characterization of an Actinobacillus equuli subsp. equuli Bearing Unique Virulence-Associated Genes from an Adult Horse with Pleuropneumonia. Pathogens 2023, 12, 224. https://doi.org/10.3390/pathogens12020224
Kamali M, Carossino M, Del Piero F, Peak L, Mitchell MS, Willette J, Baker R, Li F, Kenéz Á, Balasuriya UBR, et al. Pathological Features and Genomic Characterization of an Actinobacillus equuli subsp. equuli Bearing Unique Virulence-Associated Genes from an Adult Horse with Pleuropneumonia. Pathogens. 2023; 12(2):224. https://doi.org/10.3390/pathogens12020224
Chicago/Turabian StyleKamali, Maedeh, Mariano Carossino, Fabio Del Piero, Laura Peak, Maria S. Mitchell, Jackie Willette, Rose Baker, Fuyong Li, Ákos Kenéz, Udeni B. R. Balasuriya, and et al. 2023. "Pathological Features and Genomic Characterization of an Actinobacillus equuli subsp. equuli Bearing Unique Virulence-Associated Genes from an Adult Horse with Pleuropneumonia" Pathogens 12, no. 2: 224. https://doi.org/10.3390/pathogens12020224
APA StyleKamali, M., Carossino, M., Del Piero, F., Peak, L., Mitchell, M. S., Willette, J., Baker, R., Li, F., Kenéz, Á., Balasuriya, U. B. R., & Go, Y. Y. (2023). Pathological Features and Genomic Characterization of an Actinobacillus equuli subsp. equuli Bearing Unique Virulence-Associated Genes from an Adult Horse with Pleuropneumonia. Pathogens, 12(2), 224. https://doi.org/10.3390/pathogens12020224






