The DarT/DarG Toxin–Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies
Abstract
1. Introduction
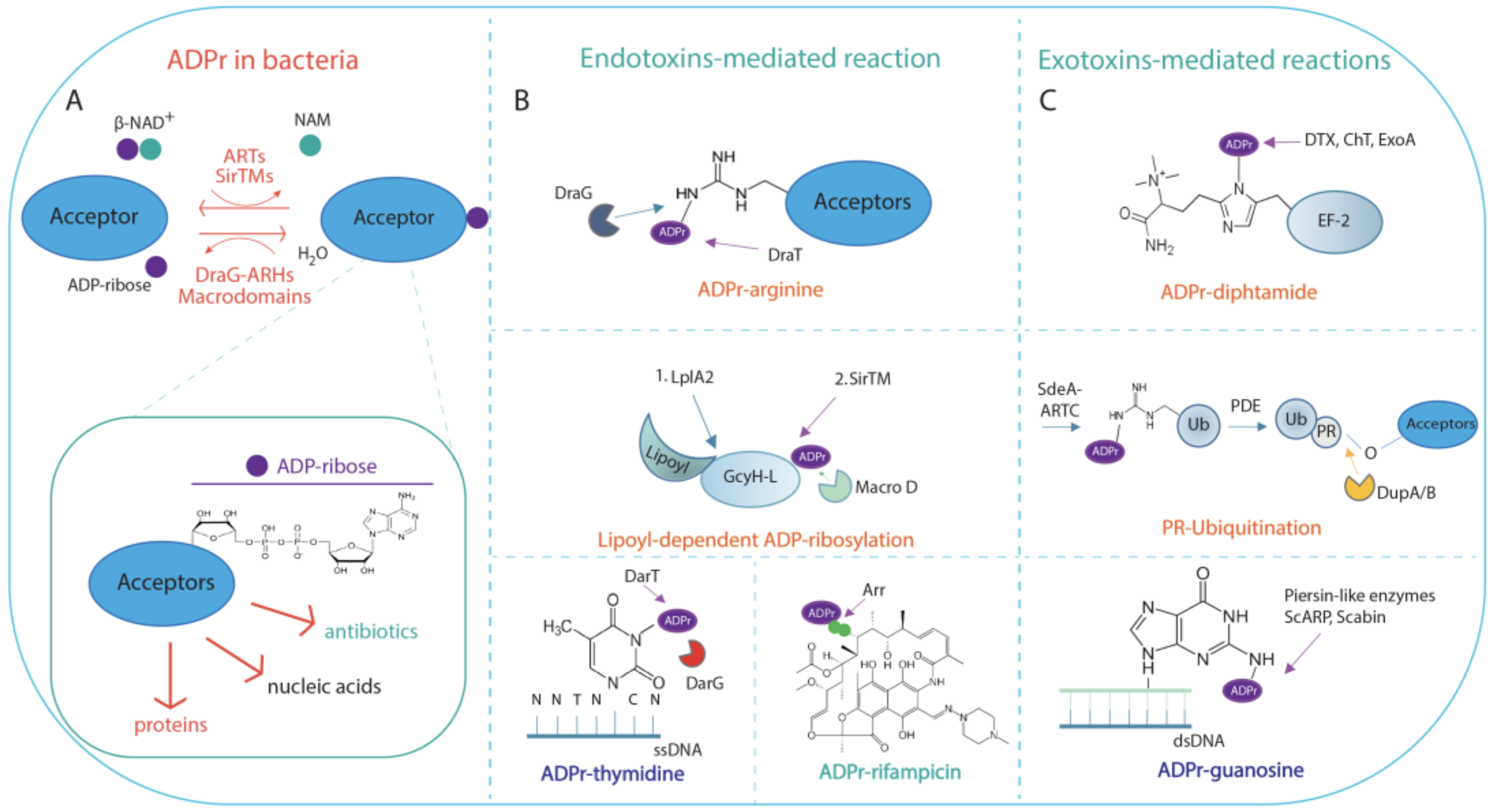
2. ADP-Ribosylation in Bacteria
2.1. NAD+-Dependent Endotoxins and Exotoxins Involved in ADP-Ribosylation Signalling
2.2. Functional Aspects of ADP-Ribosylation in Bacteria
3. The DarT/DarG ADP-Ribosylation-Dependent System
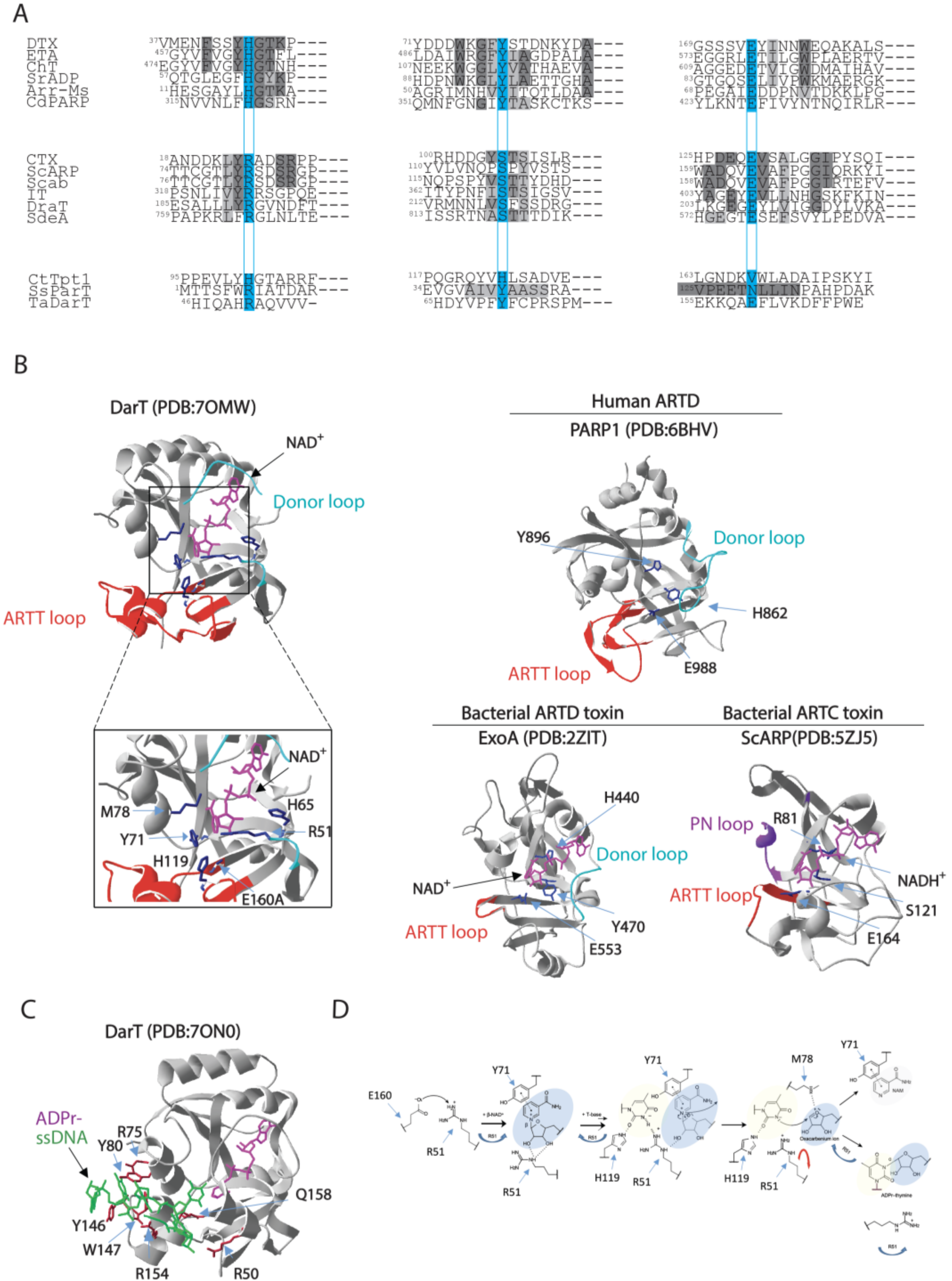
3.1. DarT Is a New PARP-Like Toxin and a Potential Molecular Target for Antimicrobial Therapy
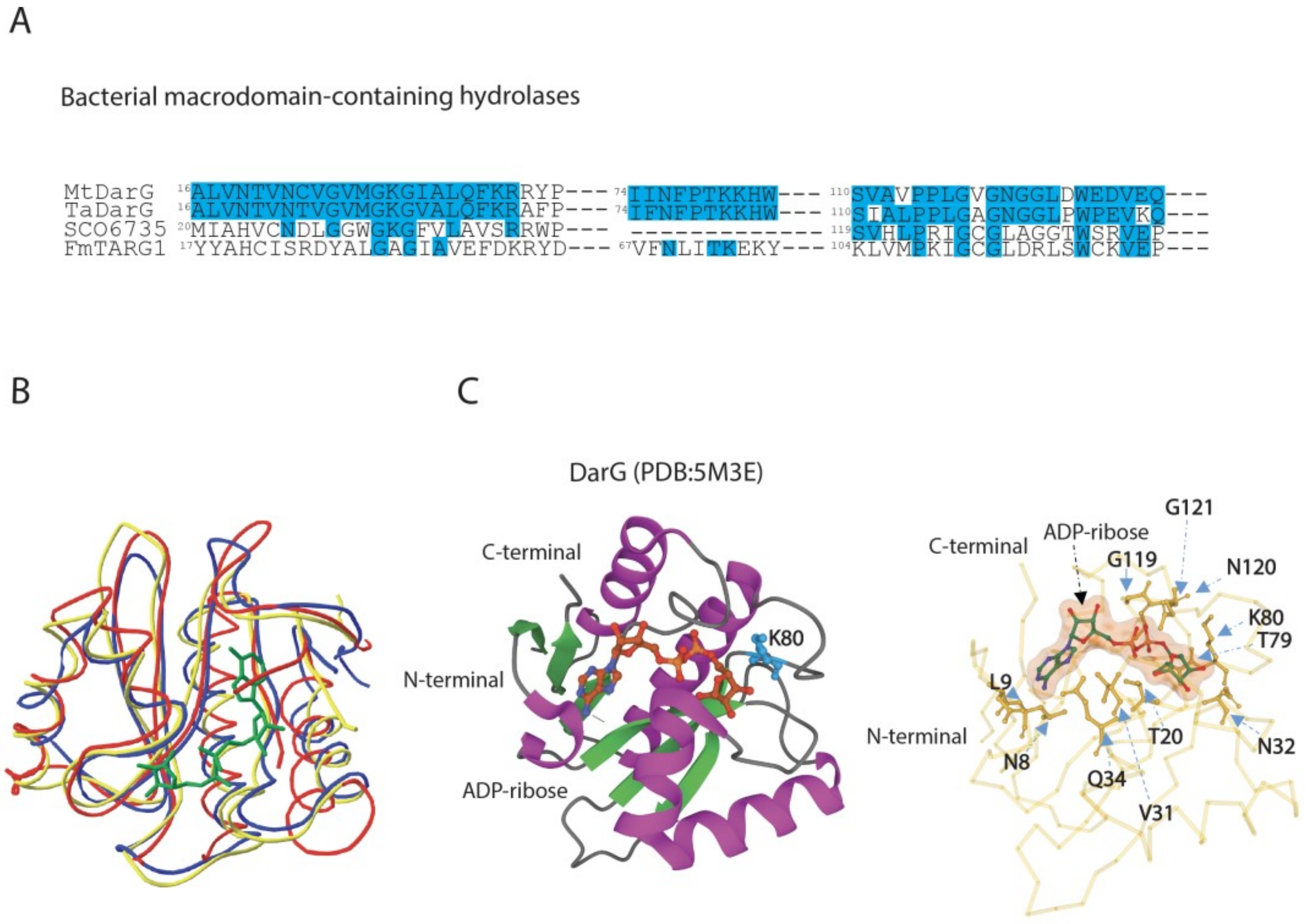
3.2. DarG Macrodomain-Containing Hydrolase Counteracts DarT Toxicity
3.3. DarT/DarG TA System Molecular Mechanisms and Biological Functions
3.4. Functional Outcomes of DarT/DarG System in Prokaryotic Immunity
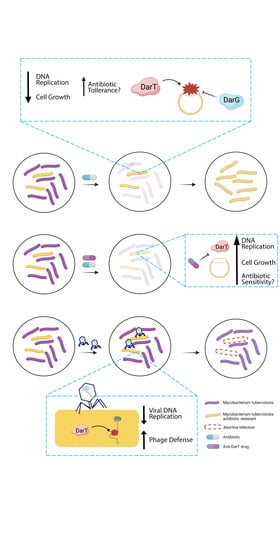
4. Exploitation of DarT/DarG Biology for a Rational Design of Antimicrobial Agents
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perina, D.; Mikoč, A.; Ahel, J.; Ćetković, H.; Žaja, R.; Ahel, I. Distribution of Protein Poly(ADP-Ribosyl)Ation Systems across All Domains of Life. DNA Repair 2014, 23, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Zhang, D.; de Souza, R.F.; Anand, S.; Iyer, L.M. The Natural History of ADP-Ribosyltransferases and the ADP-Ribosylation System. Curr. Top. Microbiol. Immunol. 2015, 384, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, L.; Mikoč, A.; Ahel, I. ADP-Ribosylation: New Facets of an Ancient Modification. FEBS J. 2017, 284, 2932–2946. [Google Scholar] [CrossRef] [PubMed]
- Mikolčević, P.; Hloušek-Kasun, A.; Ahel, I.; Mikoč, A. ADP-Ribosylation Systems in Bacteria and Viruses. Comput. Struct. Biotechnol. J. 2021, 19, 2366–2383. [Google Scholar] [CrossRef]
- Jørgensen, R.; Purdy, A.E.; Fieldhouse, R.J.; Kimber, M.S.; Bartlett, D.H.; Merrill, A.R. Cholix Toxin, a Novel ADP-Ribosylating Factor from Vibrio Cholerae. J. Biol. Chem. 2008, 283, 10671–10678. [Google Scholar] [CrossRef]
- Collier, R.J.; Pappenheimer, A.M. Studies on the mode of action of diphtheria toxin. I. Phosphorylated intermediates in normal and intoxicated hela cells. J. Exp. Med. 1964, 120, 1007–1018. [Google Scholar] [CrossRef]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-Ribosyl)Ation Reactions in the Regulation of Nuclear Functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef]
- De Vos, M.; Schreiber, V.; Dantzer, F. The Diverse Roles and Clinical Relevance of PARPs in DNA Damage Repair: Current State of the Art. Biochem. Pharmacol. 2012, 84, 137–146. [Google Scholar] [CrossRef]
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-Ribosylation: Recent Advances Linking Molecular Functions to Biological Outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef]
- Schützenhofer, K.; Rack, J.G.M.; Ahel, I. The Making and Breaking of Serine-ADP-Ribosylation in the DNA Damage Response. Front. Cell Dev. Biol. 2021, 9, 745922. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP Inhibitors: Synthetic Lethality in the Clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Palazzo, L.; Ahel, I. PARPs in Genome Stability and Signal Transduction: Implications for Cancer Therapy. Biochem. Soc. Trans. 2018, 46, 1681–1695. [Google Scholar] [CrossRef]
- Slade, D. PARP and PARG Inhibitors in Cancer Treatment. Genes Dev. 2020, 34, 360–394. [Google Scholar] [CrossRef]
- Curtin, N.J.; Szabo, C. Poly(ADP-Ribose) Polymerase Inhibition: Past, Present and Future. Nat. Rev. Drug Discov. 2020, 19, 711–736. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kraus, W.L. PARP-1 Regulates Chromatin Structure and Transcription through a KDM5B-Dependent Pathway. Mol. Cell 2010, 39, 736–749. [Google Scholar] [CrossRef]
- Gibson, B.A.; Zhang, Y.; Jiang, H.; Hussey, K.M.; Shrimp, J.H.; Lin, H.; Schwede, F.; Yu, Y.; Kraus, W.L. Chemical Genetic Discovery of PARP Targets Reveals a Role for PARP-1 in Transcription Elongation. Science 2016, 353, 45–50. [Google Scholar] [CrossRef]
- Kim, D.-S.; Camacho, C.V.; Nagari, A.; Malladi, V.S.; Challa, S.; Kraus, W.L. Activation of PARP-1 by SnoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol. Cell 2019, 75, 1270–1285.e14. [Google Scholar] [CrossRef]
- Gupte, R.; Nandu, T.; Kraus, W.L. Nuclear ADP-Ribosylation Drives IFNγ-Dependent STAT1α Enhancer Formation in Macrophages. Nat. Commun. 2021, 12, 3931. [Google Scholar] [CrossRef]
- Challa, S.; Khulpateea, B.R.; Nandu, T.; Camacho, C.V.; Ryu, K.W.; Chen, H.; Peng, Y.; Lea, J.S.; Kraus, W.L. Ribosome ADP-Ribosylation Inhibits Translation and Maintains Proteostasis in Cancers. Cell 2021, 184, 4531–4546.e26. [Google Scholar] [CrossRef]
- Jones, A.; Kraus, W.L. Multiomics Analysis of the NAD+-PARP1 Axis Reveals a Role for Site-Specific ADP-Ribosylation in Splicing in Embryonic Stem Cells. Genes Dev. 2022, 36, 601–617. [Google Scholar] [CrossRef]
- Manco, G.; Lacerra, G.; Porzio, E.; Catara, G. ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives. Biomolecules 2022, 12, 443. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.; Coughlin, M.; Mitchison, T.J. Tankyrase-1 Polymerization of Poly(ADP-Ribose) Is Required for Spindle Structure and Function. Nat. Cell Biol. 2005, 7, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Boehler, C.; Gauthier, L.R.; Mortusewicz, O.; Biard, D.S.; Saliou, J.-M.; Bresson, A.; Sanglier-Cianferani, S.; Smith, S.; Schreiber, V.; Boussin, F.; et al. Poly(ADP-Ribose) Polymerase 3 (PARP3), a Newcomer in Cellular Response to DNA Damage and Mitotic Progression. Proc. Natl. Acad. Sci. USA 2011, 108, 2783–2788. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Schreek, S.; Cerni, C.; Schamberger, C.; Lesniewicz, K.; Poreba, E.; Vervoorts, J.; Walsemann, G.; Grötzinger, J.; Kremmer, E.; et al. PARP-10, a Novel Myc-Interacting Protein with Poly(ADP-Ribose) Polymerase Activity, Inhibits Transformation. Oncogene 2005, 24, 1982–1993. [Google Scholar] [CrossRef] [PubMed]
- Andrabi, S.A.; Kang, H.C.; Haince, J.-F.; Lee, Y.-I.; Zhang, J.; Chi, Z.; West, A.B.; Koehler, R.C.; Poirier, G.G.; Dawson, T.M.; et al. Iduna Protects the Brain from Glutamate Excitotoxicity and Stroke by Interfering with Poly(ADP-Ribose) Polymer-Induced Cell Death. Nat. Med. 2011, 17, 692–699. [Google Scholar] [CrossRef]
- Bachmann, S.B.; Frommel, S.C.; Camicia, R.; Winkler, H.C.; Santoro, R.; Hassa, P.O. DTX3L and ARTD9 Inhibit IRF1 Expression and Mediate in Cooperation with ARTD8 Survival and Proliferation of Metastatic Prostate Cancer Cells. Mol. Cancer 2014, 13, 125. [Google Scholar] [CrossRef]
- Leung, A.K.L.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-Ribose) Regulates Stress Responses and MicroRNA Activity in the Cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar] [CrossRef]
- Catara, G.; Grimaldi, G.; Schembri, L.; Spano, D.; Turacchio, G.; Lo Monte, M.; Beccari, A.R.; Valente, C.; Corda, D. PARP1-Produced Poly-ADP-Ribose Causes the PARP12 Translocation to Stress Granules and Impairment of Golgi Complex Functions. Sci. Rep. 2017, 7, 14035. [Google Scholar] [CrossRef]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The Zinc-Finger Antiviral Protein Recruits the RNA Processing Exosome to Degrade the Target MRNA. Proc. Natl. Acad. Sci. USA 2007, 104, 151–156. [Google Scholar] [CrossRef]
- Atasheva, S.; Frolova, E.I.; Frolov, I. Interferon-Stimulated Poly(ADP-Ribose) Polymerases Are Potent Inhibitors of Cellular Translation and Virus Replication. J. Virol. 2014, 88, 2116–2130. [Google Scholar] [CrossRef]
- Zhang, Y.; Mao, D.; Roswit, W.T.; Jin, X.; Patel, A.C.; Patel, D.A.; Agapov, E.; Wang, Z.; Tidwell, R.M.; Atkinson, J.J.; et al. PARP9-DTX3L Ubiquitin Ligase Targets Host Histone H2BJ and Viral 3C Protease to Enhance Interferon Signaling and Control Viral Infection. Nat. Immunol. 2015, 16, 1215–1227. [Google Scholar] [CrossRef]
- Kim, C.; Wang, X.-D.; Yu, Y. PARP1 Inhibitors Trigger Innate Immunity via PARP1 Trapping-Induced DNA Damage Response. eLife 2020, 9, e60637. [Google Scholar] [CrossRef]
- Alhammad, Y.M.O.; Fehr, A.R. The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation. Viruses 2020, 12, 384. [Google Scholar] [CrossRef]
- Abd Elmageed, Z.Y.; Naura, A.S.; Errami, Y.; Zerfaoui, M. The Poly(ADP-Ribose) Polymerases (PARPs): New Roles in Intracellular Transport. Cell. Signal. 2012, 24, 1–8. [Google Scholar] [CrossRef]
- Cardamone, M.D.; Gao, Y.; Kwan, J.; Hayashi, V.; Sheeran, M.; Xu, J.; English, J.; Orofino, J.; Emili, A.; Perissi, V. Neuralized-like Protein 4 (NEURL4) Mediates ADP-Ribosylation of Mitochondrial Proteins. J. Cell Biol. 2022, 221, e202101021. [Google Scholar] [CrossRef]
- Jia, A.; Huang, S.; Song, W.; Wang, J.; Meng, Y.; Sun, Y.; Xu, L.; Laessle, H.; Jirschitzka, J.; Hou, J.; et al. TIR-Catalyzed ADP-Ribosylation Reactions Produce Signaling Molecules for Plant Immunity. Science 2022, 377, eabq8180. [Google Scholar] [CrossRef]
- Nordlund, S.; Högbom, M. ADP-Ribosylation, a Mechanism Regulating Nitrogenase Activity. FEBS J. 2013, 280, 3484–3490. [Google Scholar] [CrossRef]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef]
- Piscotta, F.J.; Jeffrey, P.D.; Link, A.J. ParST Is a Widespread Toxin-Antitoxin Module That Targets Nucleotide Metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 826–834. [Google Scholar] [CrossRef]
- Lawarée, E.; Jankevicius, G.; Cooper, C.; Ahel, I.; Uphoff, S.; Tang, C.M. DNA ADP-Ribosylation Stalls Replication and Is Reversed by RecF-Mediated Homologous Recombination and Nucleotide Excision Repair. Cell Rep. 2020, 30, 1373–1384.e4. [Google Scholar] [CrossRef]
- Kong, L.; Feng, B.; Yan, Y.; Zhang, C.; Kim, J.H.; Xu, L.; Rack, J.G.M.; Wang, Y.; Jang, J.-C.; Ahel, I.; et al. Noncanonical Mono(ADP-Ribosyl)Ation of Zinc Finger SZF Proteins Counteracts Ubiquitination for Protein Homeostasis in Plant Immunity. Mol. Cell 2021, 81, 4591–4604.e8. [Google Scholar] [CrossRef]
- Ueda, K.; Hayaishi, O. ADP-Ribosylation. Annu. Rev. Biochem. 1985, 54, 73–100. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Chang, P. Insights into the Biogenesis, Function, and Regulation of ADP-Ribosylation. Nat. Chem. Biol. 2018, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Vyas, S.; Matic, I.; Uchima, L.; Rood, J.; Zaja, R.; Hay, R.T.; Ahel, I.; Chang, P. Family-Wide Analysis of Poly(ADP-Ribose) Polymerase Activity. Nat. Commun. 2014, 5, 4426. [Google Scholar] [CrossRef] [PubMed]
- Krueger, K.M.; Barbieri, J.T. The Family of Bacterial ADP-Ribosylating Exotoxins. Clin. Microbiol. Rev. 1995, 8, 34–47. [Google Scholar] [CrossRef]
- Nakano, T.; Takahashi-Nakaguchi, A.; Yamamoto, M.; Watanabe, M. Pierisins and CARP-1: ADP-Ribosylation of DNA by ARTCs in Butterflies and Shellfish. Curr. Top. Microbiol. Immunol. 2015, 384, 127–149. [Google Scholar] [CrossRef]
- Munnur, D.; Ahel, I. Reversible Mono-ADP-Ribosylation of DNA Breaks. FEBS J. 2017, 284, 4002–4016. [Google Scholar] [CrossRef]
- Weixler, L.; Schäringer, K.; Momoh, J.; Lüscher, B.; Feijs, K.L.H.; Žaja, R. ADP-Ribosylation of RNA and DNA: From in Vitro Characterization to in Vivo Function. Nucleic Acids Res. 2021, 49, 3634–3650. [Google Scholar] [CrossRef]
- Groslambert, J.; Prokhorova, E.; Ahel, I. ADP-Ribosylation of DNA and RNA. DNA Repair 2021, 105, 103144. [Google Scholar] [CrossRef]
- Munnur, D.; Bartlett, E.; Mikolčević, P.; Kirby, I.T.; Rack, J.G.M.; Mikoč, A.; Cohen, M.S.; Ahel, I. Reversible ADP-Ribosylation of RNA. Nucleic Acids Res. 2019, 47, 5658–5669. [Google Scholar] [CrossRef]
- Baysarowich, J.; Koteva, K.; Hughes, D.W.; Ejim, L.; Griffiths, E.; Zhang, K.; Junop, M.; Wright, G.D. Rifamycin Antibiotic Resistance by ADP-Ribosylation: Structure and Diversity of Arr. Proc. Natl. Acad. Sci. USA 2008, 105, 4886–4891. [Google Scholar] [CrossRef]
- Hottiger, M.O.; Hassa, P.O.; Lüscher, B.; Schüler, H.; Koch-Nolte, F. Toward a Unified Nomenclature for Mammalian ADP-Ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Perina, D.; Ahel, I. Macrodomains: Structure, Function, Evolution, and Catalytic Activities. Annu. Rev. Biochem. 2016, 85, 431–454. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-Ribosyl)Hydrolases: Structure, Function, and Biology. Genes Dev. 2020, 34, 263–284. [Google Scholar] [CrossRef]
- Sharifi, R.; Morra, R.; Appel, C.D.; Tallis, M.; Chioza, B.; Jankevicius, G.; Simpson, M.A.; Matic, I.; Ozkan, E.; Golia, B.; et al. Deficiency of Terminal ADP-Ribose Protein Glycohydrolase TARG1/C6orf130 in Neurodegenerative Disease. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef]
- Liu, C.; Fang, Y. New Insights of Poly(ADP-Ribosylation) in Neurodegenerative Diseases: A Focus on Protein Phase Separation and Pathologic Aggregation. Biochem. Pharmacol. 2019, 167, 58–63. [Google Scholar] [CrossRef]
- Palazzo, L.; Mikolčević, P.; Mikoč, A.; Ahel, I. ADP-Ribosylation Signalling and Human Disease. Open Biol. 2019, 9, 190041. [Google Scholar] [CrossRef]
- Demény, M.A.; Virág, L. The PARP Enzyme Family and the Hallmarks of Cancer Part 1. Cell Intrinsic Hallmarks. Cancers 2021, 13, 2042. [Google Scholar] [CrossRef]
- Simon, N.C.; Aktories, K.; Barbieri, J.T. Novel Bacterial ADP-Ribosylating Toxins: Structure and Function. Nat. Rev. Microbiol. 2014, 12, 599–611. [Google Scholar] [CrossRef]
- Fehr, A.R.; Channappanavar, R.; Jankevicius, G.; Fett, C.; Zhao, J.; Athmer, J.; Meyerholz, D.K.; Ahel, I.; Perlman, S. The Conserved Coronavirus Macrodomain Promotes Virulence and Suppresses the Innate Immune Response during Severe Acute Respiratory Syndrome Coronavirus Infection. mBio 2016, 7, e01721-16. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.L.; McPherson, R.L.; Griffin, D.E. Macrodomain ADP-Ribosylhydrolase and the Pathogenesis of Infectious Diseases. PLoS Pathog. 2018, 14, e1006864. [Google Scholar] [CrossRef] [PubMed]
- Catara, G.; Corteggio, A.; Valente, C.; Grimaldi, G.; Palazzo, L. Targeting ADP-Ribosylation as an Antimicrobial Strategy. Biochem. Pharmacol. 2019, 167, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Fieldhouse, R.J.; Turgeon, Z.; White, D.; Merrill, A.R. Cholera- and Anthrax-like Toxins Are among Several New ADP-Ribosyltransferases. PLoS Comput. Biol. 2010, 6, e1001029. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Liu, C.; Shan, L.; He, P. Protein ADP-Ribosylation Takes Control in Plant-Bacterium Interactions. PLoS Pathog. 2016, 12, e1005941. [Google Scholar] [CrossRef] [PubMed]
- Toniti, W.; Yoshida, T.; Tsurumura, T.; Irikura, D.; Monma, C.; Kamata, Y.; Tsuge, H. Crystal Structure and Structure-Based Mutagenesis of Actin-Specific ADP-Ribosylating Toxin CPILE-a as Novel Enterotoxin. PLoS ONE 2017, 12, e0171278. [Google Scholar] [CrossRef]
- Belyy, A.; Lindemann, F.; Roderer, D.; Funk, J.; Bardiaux, B.; Protze, J.; Bieling, P.; Oschkinat, H.; Raunser, S. Mechanism of Threonine ADP-Ribosylation of F-Actin by a Tc Toxin. Nat. Commun. 2022, 13, 4202. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Zorzini, V.; Zhu, Z.; Schuller, M.; Ahel, D.; Ahel, I. Viral Macrodomains: A Structural and Evolutionary Assessment of the Pharmacological Potential. Open Biol. 2020, 10, 200237. [Google Scholar] [CrossRef]
- Russo, L.C.; Tomasin, R.; Matos, I.A.; Manucci, A.C.; Sowa, S.T.; Dale, K.; Caldecott, K.W.; Lehtiö, L.; Schechtman, D.; Meotti, F.C.; et al. The SARS-CoV-2 Nsp3 Macrodomain Reverses PARP9/DTX3L-Dependent ADP-Ribosylation Induced by Interferon Signaling. J. Biol. Chem. 2021, 297, 101041. [Google Scholar] [CrossRef]
- Dasovich, M.; Zhuo, J.; Goodman, J.A.; Thomas, A.; McPherson, R.L.; Jayabalan, A.K.; Busa, V.F.; Cheng, S.-J.; Murphy, B.A.; Redinger, K.R.; et al. High-Throughput Activity Assay for Screening Inhibitors of the SARS-CoV-2 Mac1 Macrodomain. ACS Chem. Biol. 2022, 17, 17–23. [Google Scholar] [CrossRef]
- Leung, A.K.L.; Griffin, D.E.; Bosch, J.; Fehr, A.R. The Conserved Macrodomain Is a Potential Therapeutic Target for Coronaviruses and Alphaviruses. Pathog. Basel Switz. 2022, 11, 94. [Google Scholar] [CrossRef]
- Roy, A.; Alhammad, Y.M.; McDonald, P.; Johnson, D.K.; Zhuo, J.; Wazir, S.; Ferraris, D.; Lehtiö, L.; Leung, A.K.L.; Fehr, A.R. Discovery of Compounds That Inhibit SARS-CoV-2 Mac1-ADP-Ribose Binding by High-Throughput Screening. Antiviral Res. 2022, 203, 105344. [Google Scholar] [CrossRef]
- Zheng, M.; Schultz, M.B.; Sinclair, D.A. NAD+ in COVID-19 and Viral Infections. Trends Immunol. 2022, 43, 283–295. [Google Scholar] [CrossRef]
- Maculins, T.; Fiskin, E.; Bhogaraju, S.; Dikic, I. Bacteria-Host Relationship: Ubiquitin Ligases as Weapons of Invasion. Cell Res. 2016, 26, 499–510. [Google Scholar] [CrossRef]
- Bhogaraju, S.; Dikic, I. Cell Biology: Ubiquitination without E1 and E2 Enzymes. Nature 2016, 533, 43–44. [Google Scholar] [CrossRef]
- Akturk, A.; Wasilko, D.J.; Wu, X.; Liu, Y.; Zhang, Y.; Qiu, J.; Luo, Z.-Q.; Reiter, K.H.; Brzovic, P.S.; Klevit, R.E.; et al. Mechanism of Phosphoribosyl-Ubiquitination Mediated by a Single Legionella Effector. Nature 2018, 557, 729–733. [Google Scholar] [CrossRef]
- Kalayil, S.; Bhogaraju, S.; Bonn, F.; Shin, D.; Liu, Y.; Gan, N.; Basquin, J.; Grumati, P.; Luo, Z.-Q.; Dikic, I. Insights into Catalysis and Function of Phosphoribosyl-Linked Serine Ubiquitination. Nature 2018, 557, 734–738. [Google Scholar] [CrossRef]
- Dikic, I.; Schulman, B.A. An Expanded Lexicon for the Ubiquitin Code. Nat. Rev. Mol. Cell Biol. 2022, 25, 1–15. [Google Scholar] [CrossRef]
- Schuller, M.; Butler, R.E.; Ariza, A.; Tromans-Coia, C.; Jankevicius, G.; Claridge, T.D.W.; Kendall, S.L.; Goh, S.; Stewart, G.R.; Ahel, I. Molecular Basis for DarT ADP-Ribosylation of a DNA Base. Nature 2021, 596, 597–602. [Google Scholar] [CrossRef]
- Rice, L.B. The Clinical Consequences of Antimicrobial Resistance. Curr. Opin. Microbiol. 2009, 12, 476–481. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- Page, R.; Peti, W. Toxin-Antitoxin Systems in Bacterial Growth Arrest and Persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Brodersen, D.E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An Overview of Toxin-Antitoxin Biology. Mol. Cell 2018, 70, 768–784. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wood, T.K. A Primary Physiological Role of Toxin/Antitoxin Systems Is Phage Inhibition. Front. Microbiol. 2020, 11, 1895. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Yadav, M.; Ghosh, C.; Rathore, J.S. Bacterial Toxin-Antitoxin Modules: Classification, Functions, and Association with Persistence. Curr. Res. Microb. Sci. 2021, 2, 100047. [Google Scholar] [CrossRef]
- Yang, Q.E.; Walsh, T.R. Toxin-Antitoxin Systems and Their Role in Disseminating and Maintaining Antimicrobial Resistance. FEMS Microbiol. Rev. 2017, 41, 343–353. [Google Scholar] [CrossRef]
- Równicki, M.; Lasek, R.; Trylska, J.; Bartosik, D. Targeting Type II Toxin–Antitoxin Systems as Antibacterial Strategies. Toxins 2020, 12, 568. [Google Scholar] [CrossRef]
- Holbourn, K.P.; Shone, C.C.; Acharya, K.R. A Family of Killer Toxins. Exploring the Mechanism of ADP-Ribosylating Toxins. FEBS J. 2006, 273, 4579–4593. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Morra, R.; Barkauskaite, E.; Kraehenbuehl, R.; Ariza, A.; Qu, Y.; Ortmayer, M.; Leidecker, O.; Cameron, D.R.; Matic, I.; et al. Identification of a Class of Protein ADP-Ribosylating Sirtuins in Microbial Pathogens. Mol. Cell 2015, 59, 309–320. [Google Scholar] [CrossRef]
- Bonfiglio, J.J.; Fontana, P.; Zhang, Q.; Colby, T.; Gibbs-Seymour, I.; Atanassov, I.; Bartlett, E.; Zaja, R.; Ahel, I.; Matic, I. Serine ADP-Ribosylation Depends on HPF1. Mol. Cell 2017, 65, 932–940.e6. [Google Scholar] [CrossRef]
- Palazzo, L.; Leidecker, O.; Prokhorova, E.; Dauben, H.; Matic, I.; Ahel, I. Serine Is the Major Residue for ADP-Ribosylation upon DNA Damage. eLife 2018, 7, e34334. [Google Scholar] [CrossRef]
- Suskiewicz, M.J.; Zobel, F.; Ogden, T.E.H.; Fontana, P.; Ariza, A.; Yang, J.-C.; Zhu, K.; Bracken, L.; Hawthorne, W.J.; Ahel, D.; et al. HPF1 Completes the PARP Active Site for DNA Damage-Induced ADP-Ribosylation. Nature 2020, 579, 598–602. [Google Scholar] [CrossRef]
- Carpusca, I.; Jank, T.; Aktories, K. Bacillus Sphaericus Mosquitocidal Toxin (MTX) and Pierisin: The Enigmatic Offspring from the Family of ADP-Ribosyltransferases. Mol. Microbiol. 2006, 62, 621–630. [Google Scholar] [CrossRef]
- Quan, S.; Venter, H.; Dabbs, E.R. Ribosylative Inactivation of Rifampin by Mycobacterium Smegmatis Is a Principal Contributor to Its Low Susceptibility to This Antibiotic. Antimicrob. Agents Chemother. 1997, 41, 2456–2460. [Google Scholar] [CrossRef]
- Ma, Y.; Ludden, P.W. Role of the Dinitrogenase Reductase Arginine 101 Residue in Dinitrogenase Reductase ADP-Ribosyltransferase Binding, NAD Binding, and Cleavage. J. Bacteriol. 2001, 183, 250–256. [Google Scholar] [CrossRef]
- Berthold, C.L.; Wang, H.; Nordlund, S.; Högbom, M. Mechanism of ADP-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-ADP-Ribosylhydrolase DraG. Proc. Natl. Acad. Sci. USA 2009, 106, 14247–14252. [Google Scholar] [CrossRef]
- Chen, D.; Vollmar, M.; Rossi, M.N.; Phillips, C.; Kraehenbuehl, R.; Slade, D.; Mehrotra, P.V.; von Delft, F.; Crosthwaite, S.K.; Gileadi, O.; et al. Identification of Macrodomain Proteins as Novel O-Acetyl-ADP-Ribose Deacetylases. J. Biol. Chem. 2011, 286, 13261–13271. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, C.; Song, Y.; Shao, C.; Zhang, X.; Zang, J. Structural Insights into the Mechanism of Escherichia Coli YmdB: A 2′-O-Acetyl-ADP-Ribose Deacetylase. J. Struct. Biol. 2015, 192, 478–486. [Google Scholar] [CrossRef]
- García-Saura, A.G.; Zapata-Pérez, R.; Hidalgo, J.F.; Sánchez-Ferrer, Á. Comparative Inhibitory Profile and Distribution of Bacterial PARPs, Using Clostridioides Difficile CD160 PARP as a Model. Sci. Rep. 2018, 8, 8056. [Google Scholar] [CrossRef]
- Cho, C.-C.; Chien, C.-Y.; Chiu, Y.-C.; Lin, M.-H.; Hsu, C.-H. Structural and Biochemical Evidence Supporting Poly ADP-Ribosylation in the Bacterium Deinococcus Radiodurans. Nat. Commun. 2019, 10, 1491. [Google Scholar] [CrossRef]
- Szirák, K.; Keserű, J.; Biró, S.; Schmelczer, I.; Barabás, G.; Penyige, A. Disruption of SCO5461 Gene Coding for a Mono-ADP-Ribosyltransferase Enzyme Produces a Conditional Pleiotropic Phenotype Affecting Morphological Differentiation and Antibiotic Production in Streptomyces Coelicolor. J. Microbiol. 2012, 50, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Lalić, J.; Posavec Marjanović, M.; Palazzo, L.; Perina, D.; Sabljić, I.; Žaja, R.; Colby, T.; Pleše, B.; Halasz, M.; Jankevicius, G.; et al. Disruption of Macrodomain Protein SCO6735 Increases Antibiotic Production in Streptomyces Coelicolor*. J. Biol. Chem. 2016, 291, 23175–23187. [Google Scholar] [CrossRef] [PubMed]
- Moure, V.R.; Costa, F.F.; Cruz, L.M.; Pedrosa, F.O.; Souza, E.M.; Li, X.-D.; Winkler, F.; Huergo, L.F. Regulation of Nitrogenase by Reversible Mono-ADP-Ribosylation. Curr. Top. Microbiol. Immunol. 2015, 384, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Prygiel, M.; Polak, M.; Mosiej, E.; Wdowiak, K.; Formińska, K.; Zasada, A.A. New Corynebacterium Species with the Potential to Produce Diphtheria Toxin. Pathog. Basel Switz. 2022, 11, 1264. [Google Scholar] [CrossRef]
- Ashida, H.; Kim, M.; Sasakawa, C. Exploitation of the Host Ubiquitin System by Human Bacterial Pathogens. Nat. Rev. Microbiol. 2014, 12, 399–413. [Google Scholar] [CrossRef]
- Qiu, J.; Luo, Z.-Q. Hijacking of the Host Ubiquitin Network by Legionella Pneumophila. Front. Cell. Infect. Microbiol. 2017, 7, 487. [Google Scholar] [CrossRef]
- Liu, Y.; Mukherjee, R.; Bonn, F.; Colby, T.; Matic, I.; Glogger, M.; Heilemann, M.; Dikic, I. Serine-Ubiquitination Regulates Golgi Morphology and the Secretory Pathway upon Legionella Infection. Cell Death Differ. 2021, 28, 2957–2969. [Google Scholar] [CrossRef]
- Schuller, M.; Ahel, I. Beyond Protein Modification: The Rise of Non-Canonical ADP-Ribosylation. Biochem. J. 2022, 479, 463–477. [Google Scholar] [CrossRef]
- Watanabe, M.; Takamura-Enya, T.; Kanazawa, T.; Totsuka, Y.; Matsushima-Hibiya, Y.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Mono(ADP-Ribosyl)Ation of DNA by Apoptosis-Inducing Protein, Pierisin. Nucleic Acids Res. Suppl. 2002, 2, 243–244. [Google Scholar] [CrossRef]
- Yamamoto, M.; Nakano, T.; Matsushima-Hibiya, Y.; Totsuka, Y.; Takahashi-Nakaguchi, A.; Matsumoto, Y.; Sugimura, T.; Wakabayashi, K. Molecular Cloning of Apoptosis-Inducing Pierisin-like Proteins, from Two Species of White Butterfly, Pieris Melete and Aporia Crataegi. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 154, 326–333. [Google Scholar] [CrossRef]
- Lyons, B.; Ravulapalli, R.; Lanoue, J.; Lugo, M.R.; Dutta, D.; Carlin, S.; Merrill, A.R. Scabin, a Novel DNA-Acting ADP-Ribosyltransferase from Streptomyces Scabies*. J. Biol. Chem. 2016, 291, 11198–11215. [Google Scholar] [CrossRef]
- Nakano, T.; Matsushima-Hibiya, Y.; Yamamoto, M.; Takahashi-Nakaguchi, A.; Fukuda, H.; Ono, M.; Takamura-Enya, T.; Kinashi, H.; Totsuka, Y. ADP-Ribosylation of Guanosine by SCO5461 Protein Secreted from Streptomyces Coelicolor. Toxicon 2013, 63, 55–63. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Zandarashvili, L.; Aguiar, P.M.; Black, B.E.; Pascal, J.M. NAD+ Analog Reveals PARP-1 Substrate-Blocking Mechanism and Allosteric Communication from Catalytic Center to DNA-Binding Domains. Nat. Commun. 2018, 9, 844. [Google Scholar] [CrossRef]
- Jørgensen, R.; Wang, Y.; Visschedyk, D.; Merrill, A.R. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008, 9, 802–809. [Google Scholar] [CrossRef]
- Yoshida, T.; Tsuge, H. Substrate N2 Atom Recognition Mechanism in Pierisin Family DNA-Targeting, Guanine-Specific ADP-Ribosyltransferase ScARP. J. Biol. Chem. 2018, 293, 13768–13774. [Google Scholar] [CrossRef]
- Talhaoui, I.; Lebedeva, N.A.; Zarkovic, G.; Saint-Pierre, C.; Kutuzov, M.M.; Sukhanova, M.V.; Matkarimov, B.T.; Gasparutto, D.; Saparbaev, M.K.; Lavrik, O.I.; et al. Poly(ADP-Ribose) Polymerases Covalently Modify Strand Break Termini in DNA Fragments in Vitro. Nucleic Acids Res. 2016, 44, 9279–9295. [Google Scholar] [CrossRef]
- Zarkovic, G.; Belousova, E.A.; Talhaoui, I.; Saint-Pierre, C.; Kutuzov, M.M.; Matkarimov, B.T.; Biard, D.; Gasparutto, D.; Lavrik, O.I.; Ishchenko, A.A. Characterization of DNA ADP-Ribosyltransferase Activities of PARP2 and PARP3: New Insights into DNA ADP-Ribosylation. Nucleic Acids Res. 2018, 46, 2417–2431. [Google Scholar] [CrossRef]
- Matta, E.; Kiribayeva, A.; Khassenov, B.; Matkarimov, B.T.; Ishchenko, A.A. Insight into DNA Substrate Specificity of PARP1-Catalysed DNA Poly(ADP-Ribosyl)Ation. Sci. Rep. 2020, 10, 3699. [Google Scholar] [CrossRef]
- Hloušek-Kasun, A.; Mikolčević, P.; Rack, J.G.M.; Tromans-Coia, C.; Schuller, M.; Jankevicius, G.; Matković, M.; Bertoša, B.; Ahel, I.; Mikoč, A. Streptomyces Coelicolor Macrodomain Hydrolase SCO6735 Cleaves Thymidine-Linked ADP-Ribosylation of DNA. Comput. Struct. Biotechnol. J. 2022, 20, 4337–4350. [Google Scholar] [CrossRef]
- Ahel, D.; Horejs, Ã.-Z.; Wiechens, N.; Polo, S.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.; Jackson, S.; et al. Poly(ADP-Ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef]
- Tromans-Coia, C.; Sanchi, A.; Moeller, G.K.; Timinszky, G.; Lopes, M.; Ahel, I. TARG1 Protects against Toxic DNA ADP-Ribosylation. Nucleic Acids Res. 2021, 49, 10477–10492. [Google Scholar] [CrossRef] [PubMed]
- Leplae, R.; Geeraerts, D.; Hallez, R.; Guglielmini, J.; Drèze, P.; Van Melderen, L. Diversity of Bacterial Type II Toxin-Antitoxin Systems: A Comprehensive Search and Functional Analysis of Novel Families. Nucleic Acids Res. 2011, 39, 5513–5525. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Poppenberger, B.; Rozhon, W. Toxin-Antitoxin Systems: Biology, Identification, and Application. Mob. Genet. Elem. 2013, 3, e26219. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Pati, S.; Kaushik, H.; Singh, S.; Garg, L.C. Toxin-Antitoxin Systems and Their Medical Applications: Current Status and Future Perspective. Appl. Microbiol. Biotechnol. 2021, 105, 1803–1821. [Google Scholar] [CrossRef] [PubMed]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and Evolution of Bacterial Toxin-Antitoxin Systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.-Y.; Rajakumar, K.; Deng, Z. TADB: A Web-Based Resource for Type 2 Toxin-Antitoxin Loci in Bacteria and Archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef]
- Akarsu, H.; Bordes, P.; Mansour, M.; Bigot, D.-J.; Genevaux, P.; Falquet, L. TASmania: A Bacterial Toxin-Antitoxin Systems Database. PLoS Comput. Biol. 2019, 15, e1006946. [Google Scholar] [CrossRef]
- Gerdes, K.; Bech, F.W.; Jørgensen, S.T.; Løbner-Olesen, A.; Rasmussen, P.B.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of Postsegregational Killing by the Hok Gene Product of the ParB System of Plasmid R1 and Its Homology with the RelF Gene Product of the E. Coli RelB Operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [CrossRef]
- Bernard, P.; Couturier, M. Cell Killing by the F Plasmid CcdB Protein Involves Poisoning of DNA-Topoisomerase II Complexes. J. Mol. Biol. 1992, 226, 735–745. [Google Scholar] [CrossRef]
- Castro-Roa, D.; Garcia-Pino, A.; De Gieter, S.; van Nuland, N.A.J.; Loris, R.; Zenkin, N. The Fic Protein Doc Uses an Inverted Substrate to Phosphorylate and Inactivate EF-Tu. Nat. Chem. Biol. 2013, 9, 811–817. [Google Scholar] [CrossRef]
- Fineran, P.C.; Blower, T.R.; Foulds, I.J.; Humphreys, D.P.; Lilley, K.S.; Salmond, G.P.C. The Phage Abortive Infection System, ToxIN, Functions as a Protein-RNA Toxin-Antitoxin Pair. Proc. Natl. Acad. Sci. USA 2009, 106, 894–899. [Google Scholar] [CrossRef]
- Masuda, H.; Tan, Q.; Awano, N.; Wu, K.-P.; Inouye, M. YeeU Enhances the Bundling of Cytoskeletal Polymers of MreB and FtsZ, Antagonizing the CbtA (YeeV) Toxicity in Escherichia Coli. Mol. Microbiol. 2012, 84, 979–989. [Google Scholar] [CrossRef]
- Wang, X.; Lord, D.M.; Cheng, H.-Y.; Osbourne, D.O.; Hong, S.H.; Sanchez-Torres, V.; Quiroga, C.; Zheng, K.; Herrmann, T.; Peti, W.; et al. A New Type V Toxin-Antitoxin System Where MRNA for Toxin GhoT Is Cleaved by Antitoxin GhoS. Nat. Chem. Biol. 2012, 8, 855–861. [Google Scholar] [CrossRef]
- Markovski, M.; Wickner, S. Preventing Bacterial Suicide: A Novel Toxin-Antitoxin Strategy. Mol. Cell 2013, 52, 611–612. [Google Scholar] [CrossRef]
- Marimon, O.; Teixeira, J.M.C.; Cordeiro, T.N.; Soo, V.W.C.; Wood, T.L.; Mayzel, M.; Amata, I.; García, J.; Morera, A.; Gay, M.; et al. An Oxygen-Sensitive Toxin-Antitoxin System. Nat. Commun. 2016, 7, 13634. [Google Scholar] [CrossRef]
- Choi, J.S.; Kim, W.; Suk, S.; Park, H.; Bak, G.; Yoon, J.; Lee, Y. The Small RNA, SdsR, Acts as a Novel Type of Toxin in Escherichia Coli. RNA Biol. 2018, 15, 1319–1335. [Google Scholar] [CrossRef]
- Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Type II Toxin-Antitoxin Systems: Evolution and Revolutions. J. Bacteriol. 2020, 202, e00763-19. [Google Scholar] [CrossRef]
- Zhang, S.-P.; Feng, H.-Z.; Wang, Q.; Kempher, M.L.; Quan, S.-W.; Tao, X.; Niu, S.; Wang, Y.; Feng, H.-Y.; He, Y.-X. Bacterial Type II Toxin-Antitoxin Systems Acting through Post-Translational Modifications. Comput. Struct. Biotechnol. J. 2021, 19, 86–93. [Google Scholar] [CrossRef]
- LeRoux, M.; Laub, M.T. Toxin-Antitoxin Systems as Phage Defense Elements. Annu. Rev. Microbiol. 2022, 76, 21–43. [Google Scholar] [CrossRef]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-Mediated Antibiotic Persistence via Phosphorylation of the Glutamyl-TRNA-Synthetase. Nat. Commun. 2013, 4, 3001. [Google Scholar] [CrossRef]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular Mechanism of Bacterial Persistence by HipA. Mol. Cell 2013, 52, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Vang Nielsen, S.; Turnbull, K.J.; Roghanian, M.; Bærentsen, R.; Semanjski, M.; Brodersen, D.E.; Macek, B.; Gerdes, K. Serine-Threonine Kinases Encoded by Split HipA Homologs Inhibit Tryptophanyl-TRNA Synthetase. mBio 2019, 10, e01138-19. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Stanger, F.V.; Scheu, P.D.; de Jong, I.G.; Goepfert, A.; Glatter, T.; Gerdes, K.; Schirmer, T.; Dehio, C. Adenylylation of Gyrase and Topo IV by FicT Toxins Disrupts Bacterial DNA Topology. Cell Rep. 2015, 12, 1497–1507. [Google Scholar] [CrossRef]
- Lu, C.; Nakayasu, E.S.; Zhang, L.-Q.; Luo, Z.-Q. Identification of Fic-1 as an Enzyme That Inhibits Bacterial DNA Replication by AMPylating GyrB, Promoting Filament Formation. Sci. Signal. 2016, 9, ra11. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Liesch, M.; Körner, J.; Québatte, M.; Engel, P.; Dehio, C. A Bacterial Toxin-Antitoxin Module Is the Origin of Inter-Bacterial and Inter-Kingdom Effectors of Bartonella. PLoS Genet. 2017, 13, e1007077. [Google Scholar] [CrossRef]
- LeRoux, M.; Srikant, S.; Teodoro, G.I.C.; Zhang, T.; Littlehale, M.L.; Doron, S.; Badiee, M.; Leung, A.K.L.; Sorek, R.; Laub, M.T. The DarTG Toxin-Antitoxin System Provides Phage Defence by ADP-Ribosylating Viral DNA. Nat. Microbiol. 2022, 7, 1028–1040. [Google Scholar] [CrossRef]
- Ting, S.-Y.; Bosch, D.E.; Mangiameli, S.M.; Radey, M.C.; Huang, S.; Park, Y.-J.; Kelly, K.A.; Filip, S.K.; Goo, Y.A.; Eng, J.K.; et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell 2018, 175, 1380–1392.e14. [Google Scholar] [CrossRef]
- Freire, D.M.; Gutierrez, C.; Garza-Garcia, A.; Grabowska, A.D.; Sala, A.J.; Ariyachaokun, K.; Panikova, T.; Beckham, K.S.H.; Colom, A.; Pogenberg, V.; et al. An NAD+ Phosphorylase Toxin Triggers Mycobacterium Tuberculosis Cell Death. Mol. Cell 2019, 73, 1282–1291.e8. [Google Scholar] [CrossRef]
- Cheverton, A.M.; Gollan, B.; Przydacz, M.; Wong, C.T.; Mylona, A.; Hare, S.A.; Helaine, S. A Salmonella Toxin Promotes Persister Formation through Acetylation of TRNA. Mol. Cell 2016, 63, 86–96. [Google Scholar] [CrossRef]
- Jurėnas, D.; Garcia-Pino, A.; Van Melderen, L. Novel Toxins from Type II Toxin-Antitoxin Systems with Acetyltransferase Activity. Plasmid 2017, 93, 30–35. [Google Scholar] [CrossRef]
- Qian, H.; Yu, H.; Li, P.; Zhu, E.; Yao, Q.; Tai, C.; Deng, Z.; Gerdes, K.; He, X.; Gan, J.; et al. Toxin-Antitoxin Operon KacAT of Klebsiella Pneumoniae Is Regulated by Conditional Cooperativity via a W-Shaped KacA-KacT Complex. Nucleic Acids Res. 2019, 47, 7690–7702. [Google Scholar] [CrossRef]
- Wilcox, B.; Osterman, I.; Serebryakova, M.; Lukyanov, D.; Komarova, E.; Gollan, B.; Morozova, N.; Wolf, Y.I.; Makarova, K.S.; Helaine, S.; et al. Escherichia Coli ItaT Is a Type II Toxin That Inhibits Translation by Acetylating Isoleucyl-TRNAIle. Nucleic Acids Res. 2018, 46, 7873–7885. [Google Scholar] [CrossRef]
- Zaveri, A.; Wang, R.; Botella, L.; Sharma, R.; Zhu, L.; Wallach, J.B.; Song, N.; Jansen, R.S.; Rhee, K.Y.; Ehrt, S.; et al. Depletion of the DarG Antitoxin in Mycobacterium Tuberculosis Triggers the DNA-Damage Response and Leads to Cell Death. Mol. Microbiol. 2020, 114, 641–652. [Google Scholar] [CrossRef]
- Magnuson, R.D. Hypothetical Functions of Toxin-Antitoxin Systems. J. Bacteriol. 2007, 189, 6089–6092. [Google Scholar] [CrossRef]
- Korch, S.B.; Hill, T.M. Ectopic Overexpression of Wild-Type and Mutant HipA Genes in Escherichia Coli: Effects on Macromolecular Synthesis and Persister Formation. J. Bacteriol. 2006, 188, 3826–3836. [Google Scholar] [CrossRef]
- Gerdes, K.; Christensen, S.K.; Løbner-Olesen, A. Prokaryotic Toxin-Antitoxin Stress Response Loci. Nat. Rev. Microbiol. 2005, 3, 371–382. [Google Scholar] [CrossRef]
- Wang, X.; Wood, T.K. Toxin-Antitoxin Systems Influence Biofilm and Persister Cell Formation and the General Stress Response. Appl. Environ. Microbiol. 2011, 77, 5577–5583. [Google Scholar] [CrossRef]
- Wen, Y.; Behiels, E.; Devreese, B. Toxin-Antitoxin Systems: Their Role in Persistence, Biofilm Formation, and Pathogenicity. Pathog. Dis. 2014, 70, 240–249. [Google Scholar] [CrossRef]
- Qiu, J.; Zhai, Y.; Wei, M.; Zheng, C.; Jiao, X. Toxin-Antitoxin Systems: Classification, Biological Roles, and Applications. Microbiol. Res. 2022, 264, 127159. [Google Scholar] [CrossRef]
- Tal, N.; Sorek, R. SnapShot: Bacterial Immunity. Cell 2022, 185, 578–578.e1. [Google Scholar] [CrossRef]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and Quantitative View of the Antiviral Arsenal of Prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef] [PubMed]
- Koga, M.; Otsuka, Y.; Lemire, S.; Yonesaki, T. Escherichia Coli RnlA and RnlB Compose a Novel Toxin-Antitoxin System. Genetics 2011, 187, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Samson, J.E.; Spinelli, S.; Cambillau, C.; Moineau, S. Structure and Activity of AbiQ, a Lactococcal Endoribonuclease Belonging to the Type III Toxin-Antitoxin System. Mol. Microbiol. 2013, 87, 756–768. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Przybilski, R.; Semeijn, K.; Salmond, G.P.C.; Fineran, P.C. A Widespread Bacteriophage Abortive Infection System Functions through a Type IV Toxin-Antitoxin Mechanism. Nucleic Acids Res. 2014, 42, 4590–4605. [Google Scholar] [CrossRef]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef]
- Saha, D.; Mukherjee, R. Ameliorating the Antimicrobial Resistance Crisis: Phage Therapy. IUBMB Life 2019, 71, 781–790. [Google Scholar] [CrossRef]
- Xu, H.-M.; Xu, W.-M.; Zhang, L. Current Status of Phage Therapy against Infectious Diseases and Potential Application beyond Infectious Diseases. Int. J. Clin. Pract. 2022, 2022, 4913146. [Google Scholar] [CrossRef]
- Årdal, C.; Balasegaram, M.; Laxminarayan, R.; McAdams, D.; Outterson, K.; Rex, J.H.; Sumpradit, N. Antibiotic Development—Economic, Regulatory and Societal Challenges. Nat. Rev. Microbiol. 2020, 18, 267–274. [Google Scholar] [CrossRef]
- Machowska, A.; Stålsby Lundborg, C. Drivers of Irrational Use of Antibiotics in Europe. Int. J. Environ. Res. Public. Health 2018, 16, 27. [Google Scholar] [CrossRef]
- Lewis, K. The Science of Antibiotic Discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.W.; Harper, D.; et al. Alternatives to Antibiotics—A Pipeline Portfolio Review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Roussin, M.; Salcedo, S.P. NAD+-Targeting by Bacteria: An Emerging Weapon in Pathogenesis. FEMS Microbiol. Rev. 2021, 45, fuab037. [Google Scholar] [CrossRef]
- Mesquita, I.; Varela, P.; Belinha, A.; Gaifem, J.; Laforge, M.; Vergnes, B.; Estaquier, J.; Silvestre, R. Exploring NAD+ Metabolism in Host-Pathogen Interactions. Cell. Mol. Life Sci. CMLS 2016, 73, 1225–1236. [Google Scholar] [CrossRef]
- van Doorn, C.L.R.; Steenbergen, S.A.M.; Walburg, K.V.; Ottenhoff, T.H.M. Pharmacological Poly (ADP-Ribose) Polymerase Inhibitors Decrease Mycobacterium Tuberculosis Survival in Human Macrophages. Front. Immunol. 2021, 12, 712021. [Google Scholar] [CrossRef]
- Lugo, M.R.; Merrill, A.R. Development of Anti-Virulence Therapeutics against Mono-ADP-Ribosyltransferase Toxins. Toxins 2020, 13, 16. [Google Scholar] [CrossRef]
- Turgeon, Z.; Jørgensen, R.; Visschedyk, D.; Edwards, P.R.; Legree, S.; McGregor, C.; Fieldhouse, R.J.; Mangroo, D.; Schapira, M.; Merrill, A.R. Newly Discovered and Characterized Antivirulence Compounds Inhibit Bacterial Mono-ADP-Ribosyltransferase Toxins. Antimicrob. Agents Chemother. 2011, 55, 983–991. [Google Scholar] [CrossRef]
- Cherubin, P.; Garcia, M.C.; Curtis, D.; Britt, C.B.T.; Craft, J.W.; Burress, H.; Berndt, C.; Reddy, S.; Guyette, J.; Zheng, T.; et al. Inhibition of Cholera Toxin and Other AB Toxins by Polyphenolic Compounds. PLoS ONE 2016, 11, e0166477. [Google Scholar] [CrossRef]
- Yates, S.P.; Taylor, P.L.; Jørgensen, R.; Ferraris, D.; Zhang, J.; Andersen, G.R.; Merrill, A.R. Structure–Function Analysis of Water-Soluble Inhibitors of the Catalytic Domain of Exotoxin A from Pseudomonas Aeruginosa. Biochem. J. 2005, 385, 667–675. [Google Scholar] [CrossRef]
- Pinto, A.F.; Ebrahimi, M.; Saleeb, M.; Forsberg, Å.; Elofsson, M.; Schüler, H. Identification of Inhibitors of Pseudomonas Aeruginosa Exotoxin-S ADP-Ribosyltransferase Activity. SLAS Discov. 2016, 21, 590–595. [Google Scholar] [CrossRef]
- Saleeb, M.; Sundin, C.; Aglar, Ö.; Pinto, A.F.; Ebrahimi, M.; Forsberg, Å.; Schüler, H.; Elofsson, M. Structure–Activity Relationships for Inhibitors of Pseudomonas Aeruginosa Exoenzyme S ADP-Ribosyltransferase Activity. Eur. J. Med. Chem. 2018, 143, 568–576. [Google Scholar] [CrossRef]
- Wu, S.-C.; Liu, F.; Zhu, K.; Shen, J.-Z. Natural Products That Target Virulence Factors in Antibiotic-Resistant Staphylococcus Aureus. J. Agric. Food Chem. 2019, 67, 13195–13211. [Google Scholar] [CrossRef]
- Abdelhamid, A.G.; El-Dougdoug, N.K. Controlling Foodborne Pathogens with Natural Antimicrobials by Biological Control and Antivirulence Strategies. Heliyon 2020, 6, e05020. [Google Scholar] [CrossRef] [PubMed]
- Bogan, A.A.; Thorn, K.S. Anatomy of Hot Spots in Protein Interfaces. J. Mol. Biol. 1998, 280, 1–9. [Google Scholar] [CrossRef]
- Modell, A.E.; Blosser, S.L.; Arora, P.S. Systematic Targeting of Protein-Protein Interactions. Trends Pharmacol. Sci. 2016, 37, 702–713. [Google Scholar] [CrossRef]
- Scott, D.E.; Bayly, A.R.; Abell, C.; Skidmore, J. Small Molecules, Big Targets: Drug Discovery Faces the Protein-Protein Interaction Challenge. Nat. Rev. Drug Discov. 2016, 15, 533–550. [Google Scholar] [CrossRef]
- Cossar, P.J.; Lewis, P.J.; McCluskey, A. Protein-Protein Interactions as Antibiotic Targets: A Medicinal Chemistry Perspective. Med. Res. Rev. 2020, 40, 469–494. [Google Scholar] [CrossRef]
- Carro, L. Protein-Protein Interactions in Bacteria: A Promising and Challenging Avenue towards the Discovery of New Antibiotics. Beilstein J. Org. Chem. 2018, 14, 2881–2896. [Google Scholar] [CrossRef]
- Kramer, V.G.; Schader, S.M.; Oliveira, M.; Colby-Germinario, S.P.; Donahue, D.A.; Singhroy, D.N.; Tressler, R.; Sloan, R.D.; Wainberg, M.A. Maraviroc and Other HIV-1 Entry Inhibitors Exhibit a Class-Specific Redistribution Effect That Results in Increased Extracellular Viral Load. Antimicrob. Agents Chemother. 2012, 56, 4154–4160. [Google Scholar] [CrossRef]
- Kahan, R.; Worm, D.J.; de Castro, G.V.; Ng, S.; Barnard, A. Modulators of Protein-Protein Interactions as Antimicrobial Agents. RSC Chem. Biol. 2021, 2, 387–409. [Google Scholar] [CrossRef]
- Gómez Borrego, J.; Torrent Burgas, M. Analysis of Host–Bacteria Protein Interactions Reveals Conserved Domains and Motifs That Mediate Fundamental Infection Pathways. Int. J. Mol. Sci. 2022, 23, 11489. [Google Scholar] [CrossRef]
- Park, S.J.; Son, W.S.; Lee, B.-J. Structural Overview of Toxin-Antitoxin Systems in Infectious Bacteria: A Target for Developing Antimicrobial Agents. Biochim. Biophys. Acta 2013, 1834, 1155–1167. [Google Scholar] [CrossRef]
- Belousova, E.A.; Ishchenko, A.A.; Lavrik, O.I. Dna Is a New Target of Parp3. Sci. Rep. 2018, 8, 4176. [Google Scholar] [CrossRef]
- Musheev, M.U.; Schomacher, L.; Basu, A.; Han, D.; Krebs, L.; Scholz, C.; Niehrs, C. Mammalian N1-Adenosine PARylation Is a Reversible DNA Modification. Nat. Commun. 2022, 13, 6138. [Google Scholar] [CrossRef]
- Fehr, A.R.; Singh, S.A.; Kerr, C.M.; Mukai, S.; Higashi, H.; Aikawa, M. The Impact of PARPs and ADP-Ribosylation on Inflammation and Host–Pathogen Interactions. Genes Dev. 2020, 34, 341–359. [Google Scholar] [CrossRef]
- Eckei, L.; Krieg, S.; Bütepage, M.; Lehmann, A.; Gross, A.; Lippok, B.; Grimm, A.R.; Kümmerer, B.M.; Rossetti, G.; Lüscher, B.; et al. The Conserved Macrodomains of the Non-Structural Proteins of Chikungunya Virus and Other Pathogenic Positive Strand RNA Viruses Function as Mono-ADP-Ribosylhydrolases. Sci. Rep. 2017, 7, 41746. [Google Scholar] [CrossRef]
- Fehr, A.R.; Jankevicius, G.; Ahel, I.; Perlman, S. Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol. 2018, 26, 598–610. [Google Scholar] [CrossRef]
- Abraham, R.; Hauer, D.; McPherson, R.L.; Utt, A.; Kirby, I.T.; Cohen, M.S.; Merits, A.; Leung, A.K.L.; Griffin, D.E. ADP-Ribosyl-Binding and Hydrolase Activities of the Alphavirus NsP3 Macrodomain Are Critical for Initiation of Virus Replication. Proc. Natl. Acad. Sci. USA 2018, 115, E10457–E10466. [Google Scholar] [CrossRef]
- Grunewald, M.E.; Chen, Y.; Kuny, C.; Maejima, T.; Lease, R.; Ferraris, D.; Aikawa, M.; Sullivan, C.S.; Perlman, S.; Fehr, A.R. The Coronavirus Macrodomain Is Required to Prevent PARP-Mediated Inhibition of Virus Replication and Enhancement of IFN Expression. PLoS Pathog. 2019, 15, e1007756. [Google Scholar] [CrossRef]
- Schuller, M.; Correy, G.J.; Gahbauer, S.; Fearon, D.; Wu, T.; Díaz, R.E.; Young, I.D.; Carvalho Martins, L.; Smith, D.H.; Schulze-Gahmen, U.; et al. Fragment Binding to the Nsp3 Macrodomain of SARS-CoV-2 Identified through Crystallographic Screening and Computational Docking. Sci. Adv. 2021, 7, eabf8711. [Google Scholar] [CrossRef]
- Gahbauer, S.; Correy, G.J.; Schuller, M.; Ferla, M.P.; Doruk, Y.U.; Rachman, M.; Wu, T.; Diolaiti, M.; Wang, S.; Neitz, R.J.; et al. Iterative Computational Design and Crystallographic Screening Identifies Potent Inhibitors Targeting the Nsp3 Macrodomain of SARS-CoV-2. BioRxiv Prepr. Serv. Biol. 2022, 120, e2212931120. [Google Scholar] [CrossRef] [PubMed]
- Sherrill, L.M.; Joya, E.E.; Walker, A.; Roy, A.; Alhammad, Y.M.; Atobatele, M.; Wazir, S.; Abbas, G.; Keane, P.; Zhuo, J.; et al. Design, Synthesis and Evaluation of Inhibitors of the SARS-CoV-2 Nsp3 Macrodomain. Bioorg. Med. Chem. 2022, 67, 116788. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Sorek, R. The Pan-Immune System of Bacteria: Antiviral Defence as a Community Resource. Nat. Rev. Microbiol. 2020, 18, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial Retrons Function In Anti-Phage Defense. Cell 2020, 183, 1551–1561.e12. [Google Scholar] [CrossRef]
- González-Delgado, A.; Mestre, M.R.; Martínez-Abarca, F.; Toro, N. Prokaryotic Reverse Transcriptases: From Retroelements to Specialized Defense Systems. FEMS Microbiol. Rev. 2021, 45, fuab025. [Google Scholar] [CrossRef]
- Bobonis, J.; Mitosch, K.; Mateus, A.; Karcher, N.; Kritikos, G.; Selkrig, J.; Zietek, M.; Monzon, V.; Pfalz, B.; Garcia-Santamarina, S.; et al. Bacterial Retrons Encode Phage-Defending Tripartite Toxin–Antitoxin Systems. Nature 2022, 609, 144–150. [Google Scholar] [CrossRef]
- Lüscher, B.; Bütepage, M.; Eckei, L.; Krieg, S.; Verheugd, P.; Shilton, B.H. ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease. Chem. Rev. 2018, 118, 1092–1136. [Google Scholar] [CrossRef]
- Iyer, L.M.; Burroughs, A.M.; Anantharaman, V.; Aravind, L. Apprehending the NAD+-ADPr-Dependent Systems in the Virus World. Viruses 2022, 14, 1977. [Google Scholar] [CrossRef]

| TA Types | Toxin | Antitoxin | Interaction Mode | Main Targets | Affected Cellular Processes | References |
|---|---|---|---|---|---|---|
| Type –I | Protein | Noncoding RNA | Interference with toxin mRNA | Bacterial membrane | Biosynthesis of cell membrane | [128] |
| Type –II | Protein | Protein | Protein–protein interaction | DNA gyrase, EF-Tu elongation factor, genomic DNA, phosphoribosyl pyrophosphate synthetase | DNA replication, translation, nucleotide metabolism | [38,39,129,130] |
| Type III | Protein | Noncoding RNA | Sequestering of the toxin | mRNA | Translation | [131] |
| Type IV | Protein | Protein | Competition for cellular targets | Cytoskeletal proteins | Cell morphology | [132] |
| Type V | RNA | Protein | Hydrolysis of toxin mRNA | Cell membrane | Biosynthesis of cell membrane | [133] |
| Type VI | Protein | Protein | Complex formation resistant to proteolysis | β-sliding clamp | DNA replication | [134] |
| Type VII | Protein | Protein | Chemical modification of the toxin | Biofilm | Biofilm | [135] |
| Type VIII | Noncoding RNA | mRNAs | Targeting of mRNAs | YhcB inner membrane protein | Cell morphology | [136] |
| Toxin | Bacterium | PTM Targets | Affected Biological Functions | References |
|---|---|---|---|---|
| HipA | E. coli K12 | Phosphorylation of Gltx | Persistence induction | [140,141] |
| HipT | E. coli O127: H6 | Phosphorylation of TrpS | Cell growth inhibition | [142] |
| Doc | E. coli | Phosphorylation of EF-Tu | Persistence induction | [130] |
| FicT | P. aeruginosa, E. coli and Yersinia enterocolitica | Adenylylation of DNA-gyrase and TopoIV | Cell growth inhibition | [143] |
| Fic-1 | P. fluorescens 2P24 | Adenylylation of DNA gyrase GyrB | Persistence induction | [144] |
| VbhT | Bartonella schoenbuchensis | T4SS effector | Secretion of virulence factors | [145] |
| DarT | M. tuberculosis | ADP-ribosylation of DNA | Cell growth inhibition Phage defence | [38,40,78,146] |
| ParT | Sphingobium sp. YBL2 | ADP-ribosylation of Prs | Cell growth inhibition | [39] |
| Tre1 | Serratia proteamaculans | ADP-ribosylation of FtsZ | Cell death | [147] |
| MbcT | M. tuberculosis | NAD+ degradation | Cell death | [148] |
| TacT | Salmonella typhimurium | Acetylation of tRNAs | Translation inhibition | [149] |
| AtaT/AtaT2 | E. coli O157:H7 | Acetylation of fMet-tRNAs | Translation inhibition | [150] |
| KacT | Klebsiella pneumoniae | Acetylation of tRNA | Translation inhibition | [151] |
| ItaT | E. coli HS | Acetylation of tRNA | Translational inhibition | [152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catara, G.; Caggiano, R.; Palazzo, L. The DarT/DarG Toxin–Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies. Pathogens 2023, 12, 240. https://doi.org/10.3390/pathogens12020240
Catara G, Caggiano R, Palazzo L. The DarT/DarG Toxin–Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies. Pathogens. 2023; 12(2):240. https://doi.org/10.3390/pathogens12020240
Chicago/Turabian StyleCatara, Giuliana, Rocco Caggiano, and Luca Palazzo. 2023. "The DarT/DarG Toxin–Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies" Pathogens 12, no. 2: 240. https://doi.org/10.3390/pathogens12020240
APA StyleCatara, G., Caggiano, R., & Palazzo, L. (2023). The DarT/DarG Toxin–Antitoxin ADP-Ribosylation System as a Novel Target for a Rational Design of Innovative Antimicrobial Strategies. Pathogens, 12(2), 240. https://doi.org/10.3390/pathogens12020240