mRNA Turnover Protein 4 Is Vital for Fungal Pathogenicity and Response to Oxidative Stress in Sclerotinia sclerotiorum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Plant Materials and Growth Conditions
2.3. Identification and Sequence Analysis of SsMRT4
2.4. Acquisition of Knockdown Mutants and Complementary Strain
2.5. Analysis of Pathogenecity
2.6. Compound Appressoria Observation and OA Analysis
2.7. RT-qPCR Analysis
2.8. DAB Staining
2.9. Abiotic Stress Response
2.10. Subcellular Localization of SsMRT4
3. Results
3.1. Identification of MRT4 in S. sclerotiorum
3.2. Knockdown and Complementation of SsMRT4 in S. sclerotiorum
3.3. SsMRT4 Is Required for Fungal Pathogenicity of S. sclerotiorum
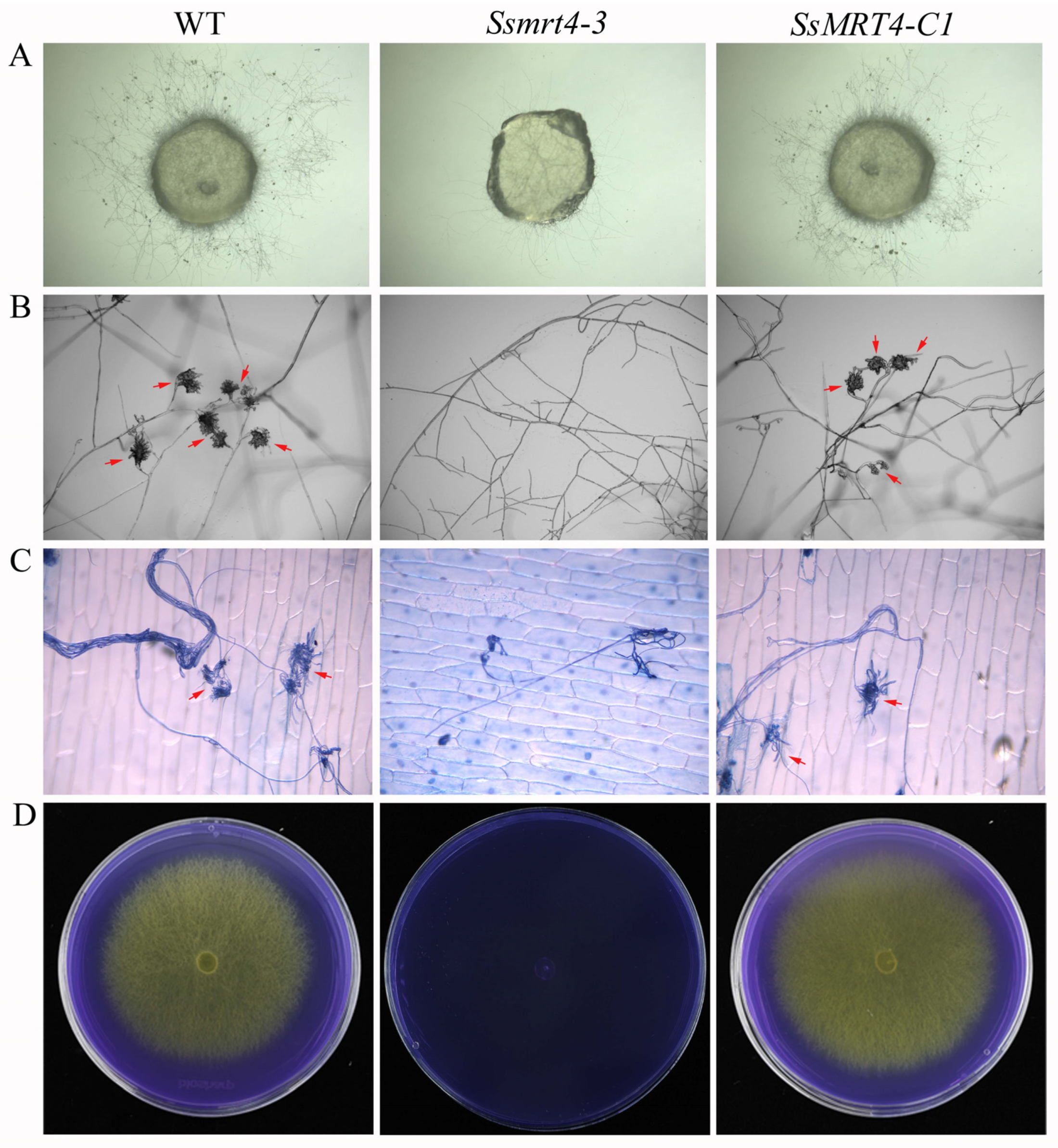
3.4. SsMRT4 Contributes to Compound Appressorium Formation and Oxalic Acid Production
3.5. SsMRT4 Is Vital for the Oxidative Stress Response
3.6. SsMRT4 Is Essential for the Cellular Integrity of Hyphae under Stresses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kressler, D.; Hurt, E.; Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2010, 1803, 673–683. [Google Scholar] [CrossRef]
- Strunk, B.S.; Karbstein, K. Powering through ribosome assembly. RNA 2009, 15, 2083–2104. [Google Scholar] [CrossRef]
- Woolford, J.L.; Baserga, S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013, 195, 643–681. [Google Scholar] [CrossRef]
- Klinge, S.; Woolford, J.L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019, 20, 116–131. [Google Scholar] [CrossRef]
- de la Cruz, J.; Katrin, K.; Woolford, J.L., Jr. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015, 84, 93–129. [Google Scholar] [CrossRef]
- Thomson, E.; Tollervey, D. Nop53p is required for late 60S ribosome subunit maturation and nuclear export in yeast. RNA 2005, 11, 1215–1224. [Google Scholar] [CrossRef]
- Fassio, C.A.; Schofield, B.J.; Seiser, R.M.; Johnson, A.W.; Lycan, D.E. Dominant mutations in the late 40S biogenesis factor Ltv1 affect cytoplasmic maturation of the small ribosomal subunit in Saccharomyces cerevisiae. Genetics 2010, 185, 199–209. [Google Scholar] [CrossRef]
- Shu, S.; Ye, K. Structural and functional analysis of ribosome assembly factor Efg1. Nucleic Acids Res. 2018, 46, 2096–2106. [Google Scholar] [CrossRef]
- Espinar-Marchena, F.; Rodriguez-Galan, O.; Fernandez-Fernandez, J.; Linnemann, J.; de la Cruz, J. Ribosomal protein L14 contributes to the early assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, 4715–4732. [Google Scholar] [CrossRef]
- Cao, S.N.; Yuan, Y.; Qin, Y.; Zhang, M.Z.; de Figueiredo, P.; Li, G.H.; Qin, Q.M. The pre-rRNA processing factor Nop53 regulates fungal development and pathogenesis via mediating production of reactive oxygen species. Environ. Microbiol. 2018, 20, 1531–1549. [Google Scholar] [CrossRef]
- Li, L.; Zhu, X.M.; Shi, H.B.; Feng, X.X.; Liu, X.H.; Lin, F.C. MoFap7, a ribosome assembly factor, is required for fungal development and plant colonization of Magnaporthe oryzae. Virulence 2019, 10, 1047–1063. [Google Scholar] [CrossRef]
- Michalec, B.; Krokowski, D.; Grela, P.; Wawiorka, L.; Sawa-Makarska, J.; Grankowski, N.; Tchorzewski, M. Subcellular localization of ribosomal P0-like protein MRT4 is determined by its N-terminal domain. Int. J. Biochem. Cell B 2010, 42, 736–748. [Google Scholar] [CrossRef]
- Rodriguez-Mateos, M.; Abia, D.; Garcia-Gomez, J.J.; Morreale, A.; de la Cruz, J.; Santos, C.; Remacha, M.; Ballesta, J.P.G. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009, 37, 3514–3521. [Google Scholar] [CrossRef]
- Sugiyama, M.; Nugroho, S.; Iida, N.; Sakai, T.; Kaneko, Y.; Harashima, S. Genetic interactions of ribosome maturation factors Yvh1 and Mrt4 influence mRNA decay, glycogen accumulation, and the expression of early meiotic genes in Saccharomyces cerevisiae. J. Biochem. 2011, 150, 103–111. [Google Scholar] [CrossRef]
- Zuk, D.; Belk, J.P.; Jacobson, A. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics 1999, 153, 35–47. [Google Scholar] [CrossRef]
- Harnpicharnchai, P.; Jakovljevic, J.; Horsey, E.; Miles, T.; Roman, J.; Rout, M.; Meagher, D.; Imai, B.; Guo, Y.; Brame, C.J.; et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 2001, 8, 505–515. [Google Scholar] [CrossRef]
- Lo, K.Y.; Li, Z.; Wang, F.; Marcotte, E.M.; Johnson, A.W. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 2009, 186, 849–862. [Google Scholar] [CrossRef]
- Rodriguez-Mateos, M.; Garcia-Gomez, J.J.; Francisco-Velilla, R.; Remacha, M.; de la Cruz, J.; Ballesta, J.P.G. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009, 37, 7519–7532. [Google Scholar] [CrossRef]
- Yang, F.; Lu, X.; Zong, H.; Ji, H.; Zhuge, B. Gene expression profiles of Candida glycerinogenes under combined heat and high-glucose stresses. J. Biosci. Bioeng. 2018, 126, 464–469. [Google Scholar] [CrossRef]
- Kakui, H.; Tsuchimatsu, T.; Yamazaki, M.; Hatakeyama, M.; Shimizu, K.K. Pollen number and ribosome gene expression altered in a genome-editing mutant of REDUCED POLLEN NUMBER1 Gene. Front. Plant Sci. 2021, 12, 768584. [Google Scholar] [CrossRef]
- Michalec-Wawiorka, B.; Wawiorka, L.; Derylo, K.; Krokowski, D.; Boguszewska, A.; Molestak, E.; Szajwaj, M.; Tchorzewski, M. Molecular behavior of human Mrt4 protein, MRTO4, in stress conditions is regulated by its C-terminal region. Int. J. Biochem. Cell Biol. 2015, 69, 233–240. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, W.; Sun, J.; Xi, Z.; Qiao, Z.; Zhang, J.; Wang, Y.; Ji, Y.; Feng, W. Screening of potential genes contributing to the macrocycle drug resistance of C. albicans via microarray analysis. Mol. Med. Rep. 2017, 16, 7527–7533. [Google Scholar] [CrossRef]
- Xia, S.; Xu, Y.; Hoy, R.; Zhang, J.; Qin, L.; Li, X. The notorious soilborne pathogenic fungus Sclerotinia sclerotiorum: An update on genes studied with mutant analysis. Pathogens 2019, 9, 27. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, J.; Lin, Y.; Fu, Y.; Xie, J.; Li, B.; Bian, X.; Feng, Y.; Liang, W.; Tang, Q.; et al. Editing homologous copies of an essential gene affords crop resistance against two cosmopolitan necrotrophic pathogens. Plant Biotechnol. J. 2021, 19, 2349–2361. [Google Scholar] [CrossRef]
- Wang, Z.; Ma, L.Y.; Cao, J.; Li, Y.L.; Ding, L.N.; Zhu, K.M.; Yang, Y.H.; Tan, X.L. Recent advances in mechanisms of plant defense to Sclerotinia sclerotiorum. Front. Plant Sci. 2019, 10, 1314. [Google Scholar] [CrossRef]
- Roper, M.; Seminara, A.; Bandi, M.M.; Cobb, A.; Dillard, H.R.; Pringle, A. Dispersal of fungal spores on a cooperatively generated wind. Proc. Natl. Acad. Sci. USA 2010, 107, 17474–17479. [Google Scholar] [CrossRef]
- Saberi-Riseh, R.; Moradi-Pour, M. A novel encapsulation of Streptomyces fulvissimus Uts22 by spray drying and its biocontrol efficiency against Gaeumannomyces graminis, the causal agent of take-all disease in wheat. Pest Manag. Sci. 2021, 77, 4357–4364. [Google Scholar] [CrossRef]
- Saberi Riseh, R.; Skorik, Y.A.; Thakur, V.K.; Moradi Pour, M.; Tamanadar, E.; Noghabi, S.S. Encapsulation of plant biocontrol bacteria with alginate as a main polymer material. Int. J. Mol. Sci. 2021, 22, 1165. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef]
- Derbyshire, M.C.; Newman, T.E.; Khentry, Y.; Owolabi Taiwo, A. The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 2022, 23, 1075–1090. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.; Belt, K.; Thatcher, L.F. Tackling control of a cosmopolitan phytopathogen: Sclerotinia. Front. Plant Sci. 2021, 12, 707509. [Google Scholar] [CrossRef]
- Luo, X.; Jiang, D.H.; Xie, J.T.; Jia, J.C.; Duan, J.; Cheng, J.S.; Fu, Y.P.; Chen, T.; Yu, X.; Li, B.; et al. genome characterization and phylogenetic analysis of a novel endornavirus that infects fungal pathogen Sclerotinia sclerotiorum. Viruses 2022, 14, 456. [Google Scholar] [CrossRef]
- Fu, M.; Pappu, H.R.; Vandemark, G.J.; Chen, W. Genome sequence of Sclerotinia sclerotiorum Hypovirulence-associated DNA Virus 1 found in the fungus Penicillium olsonii isolated from Washington State, USA. Microbiol. Resour. Announc. 2022, 11, e0001922. [Google Scholar] [CrossRef]
- Gao, Z.; Wu, J.; Jiang, D.; Xie, J.; Cheng, J.; Lin, Y. ORF Iota of Mycovirus SsNSRV-1 is associated with debilitating symptoms of Sclerotinia sclerotiorum. Viruses 2020, 12, 456. [Google Scholar] [CrossRef]
- Rollins, J.A. The Sclerotinia sclerotiorum pac1 gene is required for sclerotial development and virulence. Mol. Plant Microbe Interact. 2003, 16, 785–795. [Google Scholar] [CrossRef]
- Xu, Y.; Ao, K.; Tian, L.; Qiu, Y.; Huang, X.; Liu, X.; Hoy, R.; Zhang, Y.; Rashid, K.Y.; Xia, S.; et al. A forward genetic screen in Sclerotinia sclerotiorum revealed the transcriptional regulation of its sclerotial melanization pathway. Mol. Plant Microbe Interact. 2022, 35, 244–256. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jin, X.; Lv, Z.; Gao, J.; Zhang, R.; Zheng, T.; Yin, P.; Li, D.; Peng, L.; Cao, X.; Qin, Y.; et al. AtTrm5a catalyses 1-methylguanosine and 1-methylinosine formation on tRNAs and is important for vegetative and reproductive growth in Arabidopsis thaliana. Nucleic Acids Res. 2019, 47, 883–898. [Google Scholar] [CrossRef]
- Kabbage, M.; Yarden, O.; Dickman, M.B. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 2015, 233, 53–60. [Google Scholar] [CrossRef]
- Fan, H.; Yu, G.; Liu, Y.; Zhang, X.; Liu, J.; Zhang, Y.; Rollins, J.A.; Sun, F.; Pan, H. An atypical forkhead-containing transcription factor SsFKH1 is involved in sclerotial formation and is essential for pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 2017, 18, 963–975. [Google Scholar] [CrossRef]
- Xu, L.; Xiang, M.; White, D.; Chen, W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 2015, 17, 2896–2909. [Google Scholar] [CrossRef]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Gonzalo, P.; Reboud, J.P. The puzzling lateral flexible stalk of the ribosome. Biol. Cell 2003, 95, 179–193. [Google Scholar] [CrossRef]
- Berk, V.; Cate, J.H. Insights into protein biosynthesis from structures of bacterial ribosomes. Curr. Opin. Struct. Biol. 2007, 17, 302–309. [Google Scholar] [CrossRef]
- Rosendahl, G.; Douthwaite, S. Cooperative assembly of proteins in the ribosomal GTPase centre demonstrated by their interactions with mutant 23S rRNAs. Nucleic Acids Res. 1995, 23, 2396–2403. [Google Scholar] [CrossRef]
- Collins, S.R.; Kemmeren, P.; Zhao, X.C.; Greenblatt, J.F.; Spencer, F.; Holstege, F.C.; Weissman, J.S.; Krogan, N.J. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteom. 2007, 6, 439–450. [Google Scholar] [CrossRef]
- Schouten, A.; Van Baarlen, P.; Van Kan, J.A.L. Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associate with membranes and the nucleus of plant cells. New Phytol. 2008, 177, 493–505. [Google Scholar] [CrossRef]
- Qin, G.; Tian, S.; Chan, Z.; Li, B. Crucial role of antioxidant proteins and hydrolytic enzymes in pathogenicity of Penicillium expansum: Analysis based on proteomics approach. Mol. Cell. Proteom. 2007, 6, 425–438. [Google Scholar] [CrossRef] [Green Version]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Tang, L.; Qin, L.; Zhong, W.; Tang, X.; Gong, X.; Xie, W.; Li, Y.; Xia, S. mRNA Turnover Protein 4 Is Vital for Fungal Pathogenicity and Response to Oxidative Stress in Sclerotinia sclerotiorum. Pathogens 2023, 12, 281. https://doi.org/10.3390/pathogens12020281
Yang C, Tang L, Qin L, Zhong W, Tang X, Gong X, Xie W, Li Y, Xia S. mRNA Turnover Protein 4 Is Vital for Fungal Pathogenicity and Response to Oxidative Stress in Sclerotinia sclerotiorum. Pathogens. 2023; 12(2):281. https://doi.org/10.3390/pathogens12020281
Chicago/Turabian StyleYang, Chenghuizi, Lan Tang, Lei Qin, Weiping Zhong, Xianyu Tang, Xin Gong, Wenqi Xie, Yifu Li, and Shitou Xia. 2023. "mRNA Turnover Protein 4 Is Vital for Fungal Pathogenicity and Response to Oxidative Stress in Sclerotinia sclerotiorum" Pathogens 12, no. 2: 281. https://doi.org/10.3390/pathogens12020281
APA StyleYang, C., Tang, L., Qin, L., Zhong, W., Tang, X., Gong, X., Xie, W., Li, Y., & Xia, S. (2023). mRNA Turnover Protein 4 Is Vital for Fungal Pathogenicity and Response to Oxidative Stress in Sclerotinia sclerotiorum. Pathogens, 12(2), 281. https://doi.org/10.3390/pathogens12020281





