Abstract
Theobroma cacao is one of the main crops of economic importance in the world as the source of raw material for producing chocolate and derivatives. The crop is the main source of income for thousands of small farmers, who produce more than 80% of the world’s cocoa supply. However, the emergence, re-emergence and proliferation of pathogens, such as Ceratocystis spp., the causative agent of Ceratocystis wilt disease and canker disease, have been affecting the sustainability of many crops. Fungal control is laborious, often depending on fungicides that are expensive and/or toxic to humans, prompting researchers to look for new solutions to counteract the proliferation of these pathogens, including the use of biological agents such as mycoviruses. In this study, we investigated the diversity of microorganisms associated with the T. cacao pathogens Ceratocystis cacaofunesta and Ceratocystis fimbriata with a focus on the virome using RNA sequencing data available in public databases. We used a comprehensive bioinformatics pipeline containing several steps for viral sequence enrichment and took advantage of an integrated assembly step composed of different assemblers followed by sequence similarity searches using NCBI nonredundant databases. Our strategy was able to identify four putative C. cacaofunesta viruses (hypovirus, sclerotimonavirus, alphapartitivirus and narnavirus) and six C. fimbriata viruses (three alphaendornaviruses, one victorivirus and two mitoviruses). All the viral sequences identified showed similarity to viral genomes in public databases only at the amino acid level, likely representing new viral species. Of note, we present the first report of viruses associated with the cacao pathogens C. cacaofunesta and C. fimbriata and the second report of viral species infecting members of the Ceratocystidaceae family. Our findings highlight the need for further prospective studies to uncover the real diversity of fungus-infecting viruses that can contribute to the development of new management strategies.
1. Introduction
Theobroma cacao L. is one of the most important cash crops in many tropical regions of the world, with planted areas comprising ~8.2 million ha [1]. The nuts are used to make chocolate and cosmetics [2]. The cacao crop is the main source of income for millions of small farmers in many countries (e.g., Ivory Coast, Ghana, Ecuador and Brazil), who account for more than 80% of the global supply of cocoa [3,4]. Overall, worldwide food security is threatened by the effects of biotic and abiotic stresses on crops, with environmental (mostly climatic) and genetic factors being the main causes of new pathogens and insect pests. Under these circumstances, a major challenge for plant breeders is the generation and improvement of resistant crops [5], since phytopathogens can reduce yields by up to 20% or more [6].
Ceratocystis spp. (Ascomycota phylum, Sordariomycetes class) include aggressive pathogenic fungal species, which attack an array of economically important plant species worldwide. These pathogens cause the diseases Ceratocystis wilt and canker, which often kill the host plants, causing significant losses to farmers and jeopardizing the sustainability of agricultural production [7]. In the last two decades, emerging Ceratocystis diseases have increased considerably in Central and South America [7,8]; Theobroma cacao L. and Eucalyptus sp. can be infected by Ceratocystis cacaofunesta and C. fimbriata, respectively, which reduces the productive capacity of these plants [9]. The causal agent of Ceratocystis wilt of cacao (CWC, or ‘machete disease’) [10] is one of the three well-established host-specific pathogens in the genus [7], causing disease only to cacao plants [11]. Besides pruning, another transmission mechanism of this pathogen is through contact of the plant with different insect vectors, such as those of the Xyleborus genus (Coleoptera-Scolytidae) [10,12,13]. This fungus penetrates the plant through holes and wounds. The CWC pathogenesis is characterized by irreversible damage to the plant’s vascular system, leading to death on the order of 104 to 105 individuals per plantation, hence significantly reducing cocoa production per area [7,14]. This disease poses a serious threat to global cocoa production due to the high risk of its spread [7,15] by human activities, the main propagation mode described [3].
The control of this fungal pathogen is difficult, so the search for alternative economically viable and environmentally friendly solutions is necessary [14]. Previous research has described the potential of controlling oak wilt (caused by C. fimbriata) by using the bacterium Pseudomonas chlororaphis subsp. aureofaciens, based on the synthesis of volatile compounds by SPS-41, which showed significant inhibitory effects on mycelial growth and spore germination of C. fimbriata [16]. In terms of cacao (a perennial plant), one of the most efficient strategies to minimize losses is the selection of resistant varieties. However, the substitution of existing genotypes with resistant genotypes on plantations is a slow process. In addition, the various cacao pathogens and their specific modes of pathogenicity hamper the selection of plants simultaneously resistant to all of them [2]. Each pathogen affects different organs of the plant, e.g., Moniliophthora perniciosa in vegetative shoots, pods, and floral cushions [17]; Phytophthora spp. in pods and leaves; [18] and Ceratocystis cacaofunesta in the vascular system [11]. Finally, the use of biocontrol agents (BCAs) such as Trichoderma spp. has proven to be effective in inhibiting the in vitro growth and reducing spore germination of C. cacaofunesta [19]. The antifungal tebuconazole present in the medicinal plant Adiantum latifolium also had an important inhibition effect on mycelial growth and propagation of C. cacaofunesta, mainly in places where genetic selection and proper handling of instruments at the time of pruning fail altogether [14].
Among the BCAs, mycoviruses are good candidates, since they have been found to infect all major taxonomic groups of fungi, including phytopathogenic species [20]. Several mycoviruses cause abnormal symptoms in the host, such as hypovirulence and wasting [21]. A successful example of a viral BCA against phytopathogenic fungi is Cryphonectria hypovirus 1 (CHV1), which causes hypovirulence in Cryphonectria parasitica and has been used to reduce infection of figs [22]. Similarly, Fusarium oxysporum alternavirus 1 (FoAV1) was recently found to reduce the fungal growth and symptoms of Fusarium wilt (a vascular disease) in Lilium brownii [23]. For phytopathogenic fungi such as Sclerotinia sclerotiorum, S. nivalis, S. minor and Botrytis cinerea, Sclerotinia sclerotiorum partitivirus 1 can affect the virulence of the host fungus on the plant [24]. Thus, these and other mycovirus species are very promising as biological control agents against fungal diseases [21,25,26].
Fungi have developed different defense mechanisms against their own pathogens, including viruses. The conserved defense mechanisms based on RNA interference (RNAi) viral silencing have been studied in many fungal species, including Cryponectria parasitica, Aspergillus nidulans and Rosellinia necatrix [27,28]. These mechanisms act against invading nucleic acids, such as those produced by viruses, transposons or transgenes, in a wide spectrum of fungal species [26,29,30,31]. In fungi, the different RNAi pathways are related to three main functions: genomic defense, heterochromatin formation and gene regulation [32,33,34,35]. Although most fungal species possess the RNAi mechanism, there are several filamentous fungi (e.g., Candida albicans and Ustilago maydis [27,36]) and yeasts (e.g., Saccharomyces cerevisiae [32]) that seem to lack all or most of the components necessary for RNA silencing to occur [27,36]. This means that the RNAi pathway is not required by some unicellular organisms [32]. Fungal viruses are less explored than plant viruses, therefore their real diversity still unknown. Similarly, regarding plant viruses, most mycoviruses have been studied in the context of controlling fungal phytopathogens [37]. The application of bioinformatics and next-generation sequencing technologies (NGS) has contributed greatly to the discovery of new viruses in many organisms, including fungi [26]. NGS techniques have been used to detect the presence of viral sequences, regardless of the viral titer of the sample, since this method does not require prior knowledge of the genomic sequences of the candidate viruses [26,37,38].
We decided to investigate the virome associated with the important T. cacao pathogens C. cacaofunesta and C. fimbriata using public RNA sequencing data. Our bioinformatics strategy led to the identification of many viruses in both Ceratocysitis species, including members of Hypoviridae, Alphaendornaviridae and Alphapartitiviridae families that have been described as containing viruses with biotechnological potential. Of note, this is the first report of viruses associated with the cacao pathogens C. cacaofunesta and C. fimbriata and the second report of viruses infecting members of the Ceratocystidaceae family. Our findings highlight the need for new studies to uncover the real diversity of mycoviruses that can contribute to the development of new biofungicides.
2. Materials and Methods
2.1. Acquisition of RNA Libraries
Ceratocystis cacaofunesta and Ceratocystis fimbriata public RNA-seq libraries were downloaded from the Sequence Read Archive (SRA) database from NCBI. We used two libraries: one from C. cacaofunesta (SRR6217952) constructed from a medium enriched with cacao xylem and deeply sequenced using the Illumina HiSeq 2500 Platform with a single layout [10] and one from C. fimbriata isolate CBS 114,723 (SRR8599076) constructed from a culture maintained at the Forestry and Agricultural Biotechnology Institute (FABI) of the University of Pretoria, which was deeply sequenced using the Ion Torrent Proton instrument, also with a single layout [39,40].
2.2. Bioinformatic Analysis
2.2.1. Transcriptome Assembly
Public RNA-seq libraries were preprocessed using the tools FastQC (version 0.11.9) [41] and Trim Galore (version 0.6.7 + galaxy0) [42]. Removal of host-derived sequences was performed by aligning preprocessed reads against each specific reference genome—C. cacaofunesta (PEJQ01000008.1) and C. fimbriata (VNIJ01000001.1)—using Bowtie2 (version 2.4.5 + galaxy1) [43] with default settings. The unmapped reads were used in the assembly step, taking advantage of different assemblers: SPAdes (version 3.15.4 + galaxy1) [44], rnaviralSPAdes (version 3.15.4 + galaxy1) [45] and Trinity (version 2.9.1 + galaxy2) [46], with default settings. The assembled transcripts derived from SPAdes, rnaviralSPAdes and Trinity were used as input for CAP3 (version 10.2011) [47] in order to consolidate the results and extend possible viral sequences.
2.2.2. Identification of Viral Sequences
The viral contigs were identified by sequence similarity search through the Diamond (version 2.0.15 + galaxy0) [48] with BlastX mode using the viral Refseq from NCBI as the database. Statistical analysis of assembled contigs was performed with Fasta Statistics (version 2.0) [49] (Supplementary Table S1). All tools and programs used can be found online on the Galaxy Australia platform [50].
2.2.3. Manual Curation of Viral Genomes
Non-retroviral sequences were filtered by length >500 nt and checked by manual inspection including online Blast searches, structural annotation, and analysis of conserved domains. Nonredundant contigs were subjected to sequence similarity searches using the online Basic Local Alignment Search Tool [51] at the nucleotide and amino acid levels. Sequences identified with coverage greater than 30% with known species and E-values < 1 × 10−3 for protein and <1 × 10−5 for nucleotide were further analyzed (Supplementary Table S2). Representative viral sequences from each group were selected, classified according to the highest bit score and used in subsequent analyses. Structural annotation regarding predicted open reading frames (ORFs) was carried out using the online tool ORFfinder [52] except for viruses related to the Mitoviridae and Narnaviridae families, for which genetic code 4 was used (Mold, Mitochondrial Protozoa and Coelenterates and Mycoplasma/Spiroplasma), with acceptance of “ATG” and alternative initiation codons. The ORFs were used for the identification of conserved protein domains as well as homologous superfamilies of the different amino acids of replication-associated proteins in the viruses C. cacaofunesta and C. fimbriata using the online tools InterProScan [53] and HMMER [54,55,56].
2.2.4. Integrative Genome Assembly
In order to improve possible viral genomes, an integrative assembly strategy was also performed running the different tools rnaviralSPAdes [45], metaSPAdes [57], Oases [58], Metavelvet [59] and IDBA_UB [60]. The results were consolidated using CD-HIT [61] to join sequences with more than 90% similarity. The assembled sequences were manually investigated and used along with raw read alignment to correct misassembled regions. Manually curated viral sequences identified in our work were deposited in the NCBI GenBank Third Party Annotation database under accession numbers BK062942-BK062946, BK063053-BK063059, and can also be found in the Supplementary Data.
2.3. Phylogenetic Analysis
Viral sequences that presented similarity to polymerase or polyprotein-coding genes were used in phylogenetic analysis. Datasets composed of public protein sequences (Supplementary Table S3) related to Hypoviridae, Fusariviridae, Nyamiviridae, Mymonaviridae, Potyviridae, Partitiviridae, Fiersviridae, Botourmiaviridae, Narnaviridae, Mitoviridae, Tymoviridae, Endornaviridae, Cystoviridae and Totiviridae were created considering close amino acid viral sequences and ICTV reference sequences. For each dataset, a global alignment was created using the online tool MAFFT [62]. The alignment was visualized and trimmed with AliView [63]. The edited alignment was used as input in the program ModelTest to select the best evolutionary amino acid model according to the Akaike information criterion (AIC), and then a maximum likelihood phylogeny tree was inferred with 1000 bootstrap replicates. This last step was performed with the CIPRES Science Gateway [64]. The trees were visualized in the online tool Interactive Tree of Life (ITOL v6) [65]. Lastly, to search for conserved RNA-dependent RNA polymerase (RdRp) motifs within the putative viral proteins, we used the NCBI CDD, v3.20-59693 PSSM search tool with standard parameters [66].
2.4. Genetic Diversity of Species and Abundance of Viral Sequences
The assembled consolidated transcripts derived from SPAdes, rnaviralSPAdes and Trinity assemblies were analyzed using the Kaiju platform [67] with standard parameters (Supplementary Table S4). In addition, to produce the reference transcriptome, we performed a second contig assembly using the programs SPAdes (version 3.15.4 + galaxy1) [44], rnaviralSPAdes (version 3.15.4 + galaxy1) [45] and Trinity (version 2.9.1 + galaxy2) [46], with raw reads followed by TransDecoder (version 5.5.0 + galaxy2) [68] to identify high-confidence transcripts. Finally, the Salmon quant software (version 1.5.1 + galaxy0) [69] was executed using the identified transcripts and viral sequences to assess viral abundance. The host mitochondrial ribosomal protein S3 and nuclear beta-tubulin genes were selected as endogenous and standard genes for comparison with viral quantification.
2.5. Characterization of RNAi-Related Proteins in C. fimbriata
Identification: The proteins involved in the RNAi pathway, Dicer proteins 1 and 2 (DCL1 and DCL1), RNase III domain-containing protein (MRPL3), quelling deficient element genes (QDE), QDE-2-interacting protein (QIP), suppressor of ascus dominance (SAD) and suppressor of meiotic silencing (SMS) were identified based on annotation derived from Neurospora crassa [32]. The N. crassa proteins were aligned against the Ceratocystis fimbriata annotation (Cfim3.0 NCBI) using sequence similarity searches with the Blast software. The homologous proteins identified were evaluated for the presence of conserved domain characteristics of each class of proteins using the InterProScan 5.59–91.0 tool. Dendrogram: Homologous proteins from N. crassa and C. fimbriata were aligned by the ClustalW tool [70], and analyzed with the parameters of distance calculation, fractional dissimilarity and bootstrap value of 100. The location and name of the identified domains of each sequence were used to construct the domain plot of the dendrogram with the Archeopteryx 0.9914 tool [71]. Abundance: Transcripts corresponding to RNAi-related proteins in C. fimbriata were identified in their corresponding genome (Cfim3.0 NCBI) and used as a reference for quantification. The C. fimbriata transcriptome library (SRR16204154) was used for quantification, using the Salmon quant tool of the Galaxy platform with standard parameters.
3. Results
3.1. Transcriptome Assembly
The C. cacaofunesta (Cc) library presented 55,951,393 raw reads. After the quality filtering, 55,529,085 reads with Phred quality >20 were retained for further analysis. A total of 91.15% of the reads were aligned with the fungal reference genome. The reads that were not aligned with the reference genome were used in the transcriptome assembly. The assembly produced 1750 SPAdes transcripts, 965 rnaviralSPAdes transcripts and 480 Trinity transcripts. The consolidation and transcript extension step with CAP3 resulted in 1002 unambiguous sequences with N50 of 1932 and an average length of 892.9 nt. The C. fimbriata (Cf) library consisted of 42,480,313 raw reads. A total of 68.55% of the reads were aligned with the reference genome. The remaining reads were used in the assembly step, producing 7398 SPAdes transcripts, 10,709 rnaviralSPAdes transcripts and 64,961 Trinity transcripts. Transcript consolidation resulted in 52,493 nonredundant sequences with N50 of 729 and an average length of 593 nt (Supplementary Table S1).
3.2. Metagenomic Analysis
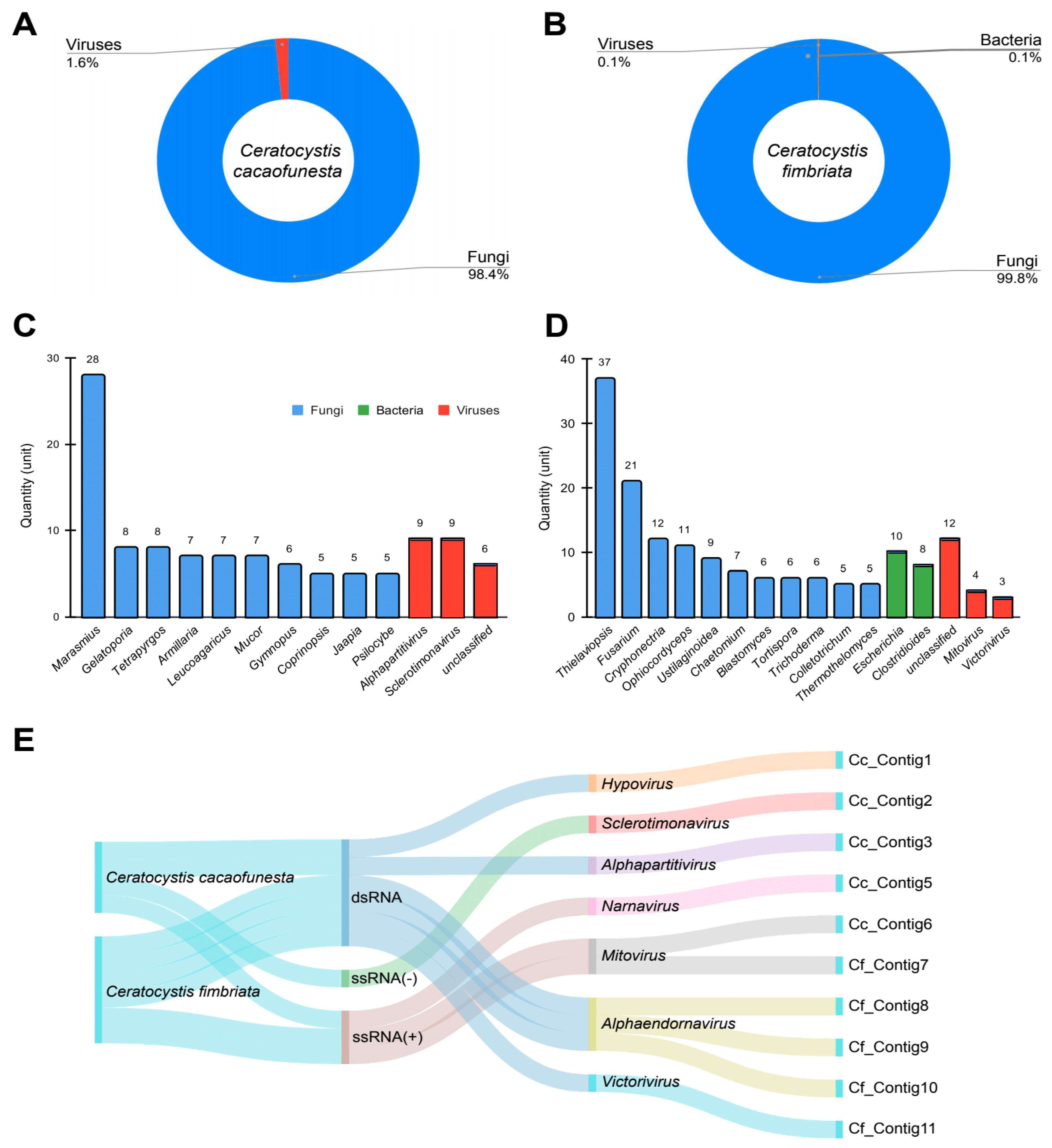
Transcriptomic data resulting from the consolidation of the data from the three different assemblers were used for initial screening using a metagenomics-based strategy with Kaiju. We observed transcripts derived mainly from fungi (Cc: 98.4%, Cf: 99.8%) and viruses (Cc: 1.6%, Cf: 0.1%) in Ceratocystis samples, with the exception of C. fimbriata, in which we also observed sequences derived from bacteria (0.1%) (Figure 1A,B). Regarding fungal diversity, in the sample derived from C. cacaofunesta, we observed the presence of transcripts assigned to species from different genera, such as Marasmius, Gelatoporia and Armilaria, while in C. fimbriata, we detected sequences related to Thielaviopsis, Fusarium, Trichoderma and Colletotrichum, among others (Figure 1C,D). The viral sequences detected included members of alphapartitivirus, sclerotimonavirus, an unclassified genus in C. cacaofunesta and mitovirus, victorivirus and an unclassified genus in C. fimbriata (Figure 1C,D). Sequences derived from bacteria were only identified in C. fimbriata and were restricted to species related to Escherichia and Clostridioides (Figure 1D).
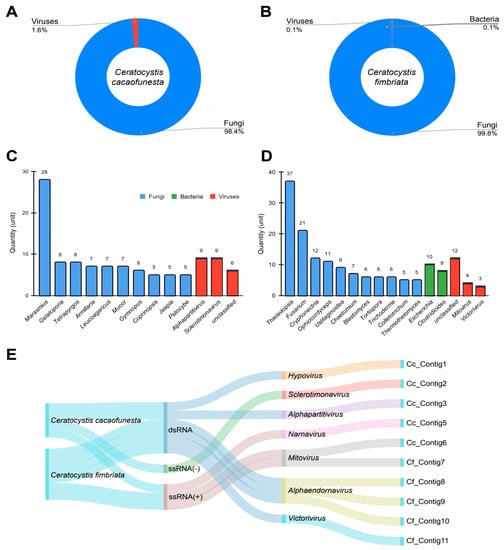
Figure 1.
Metagenomic analysis of Ceratocystis samples. Diversity of microorganisms identified in C. cacaofunesta and C. fimbriata at the kingdom (A,B) and genus (C,D) level, respectively. (E) Sankey plot referring to the general diversity of transcripts derived from viruses identified in Ceratocystis samples classified by genome composition and viral family.
3.3. Virome Characterization
We identified a considerable number of viral sequences in the metagenomicanalysis, so we further explored the presence of viruses in Ceratocystis samples. Therefore, we performed regular sequence similarity searches against NCBI nonredundant databases. For C. cacaofunesta and C. fimbriata, viral analysis identified the presence of double-stranded RNA viruses (dsRNA) and positive single-stranded RNA viruses (ssRNA+), while only in the case of C. cacaofunesta, we also identified elements relative to negative single-stranded RNA viruses (ssRNA−) (Figure 1E). At the genus level, members of the dsRNA genome included hypovirus, alphapartitivirus, alphaendornavirus and victorivirus, while ssRNA(+) was represented by narnavirus and mitovirus and, finally, ssRNA(−) by sclerotimonavirus (Figure 1E).
- Hypoviridae
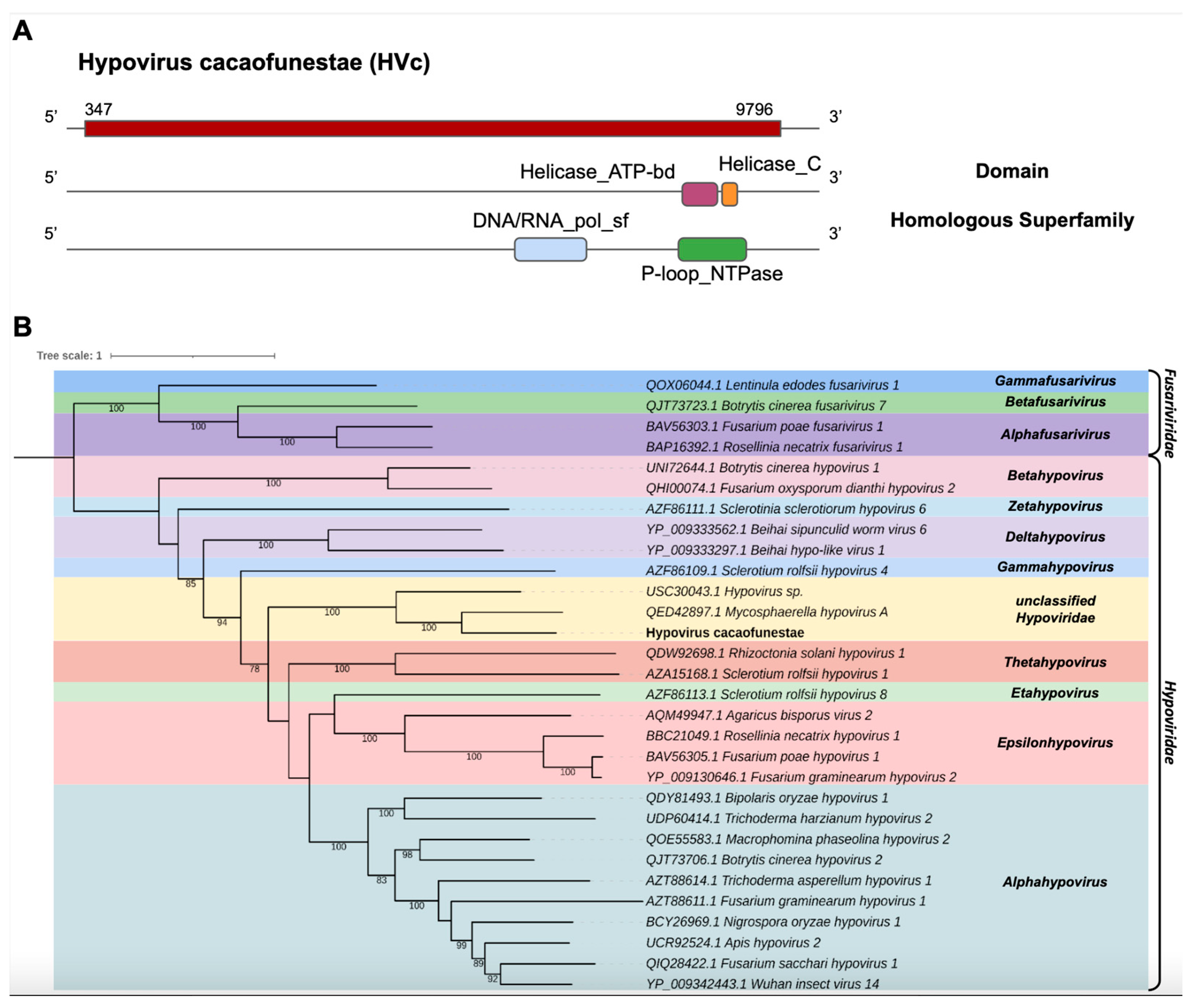
The transcript assembled from the library of C. cacaofunesta, Cc_Contig1, presented 9920 nt and had similarity to viruses of the family Hypoviridae at the amino acid level. Structural annotation using NCBI ORFfinder indicated a large open reading frame (ORF) of 9450 nt surrounded by UTRs of 5′ and 3′ with 347 nt and 124 nt, respectively (Figure 2A). In Cc_Contig1, we detected the domains Helicase_ATP-bd (IPR014001) and Helicase_C (IPR001650) superimposed on the homologous superfamily P-loop_NTPase (IPR027417) and the DNA/ARN_pol_sf superfamily (IPR043502) (Figure 2A). According to the phylogenetic analysis, the putative virus was grouped with sequences from the genus Hypovirus (Hypoviridae), closely related to Mycosphaerella hypovirus A (MHV-A), with 100% bootstrap (Figure 2B). Indeed, analysis of conserved domains of the closely related MHV-A revealed the same domains identified in the Cc_Contig1, with the exception of the RNA-dir_pol_PSvirus domain (IPR007094), only identified in the MHV-A (Figure 2A and Supplementary Figure S1). This new virus was named Hypovirus cacaofunestae (HVc) to reflect the host origin.
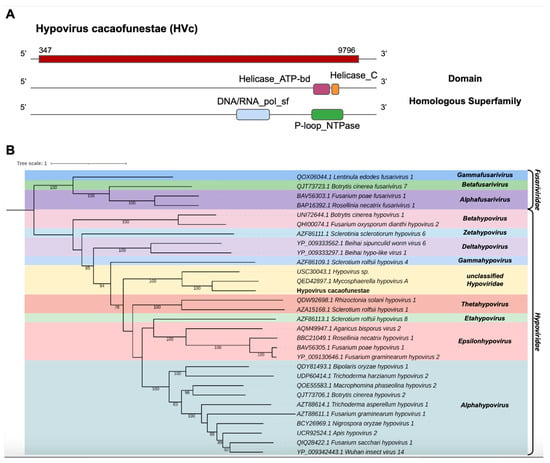
Figure 2.
Characterization of the new Hypovirus. (A) Schematic representation of the novel hypovirus HVc with the ORF and domains represented. (B) Phylogenetic analysis using the ModelTest-NG according to the Akaike information criterion (AIC) indicated that the best evolutionary model was BLOSUM62 + F. Bootstrap values were established from 1000 replicates. Bootstrap values (%) lower than 70 are not shown.
Of note, in the HVc viral contig, we also identified conserved domains (CDD) that presented a central catalytic domain of RdRp of RNA viruses belonging to the Hypoviridae family, ps-ssRNAv_Hypoviridae_RdRp (cd23170), which is a member of the ps-ssRNAv_RdRp-like superfamily (cl40470) located in the interval 2079–23038 and with an E-value of 1.53 × 10−20. This region represents conserved polymerase motifs called conserved polymerase motif A, located at position 2149–2163 TRFSADITAYDANTP; polymerase B motif, located at positions 2217–2233 and 2235–2240 GGTGQSSTSWDNHWGMR..AMIMIW; and conserved polymerase C motif, located at positions 2255–2258 and 2259–2269 NSVH..TGDDNIWGTD, sites that are found in seven (there are only six identified) viral species, namely, Cryphonectria hypovirus 3 (AAF13604), Phomopsis longicolla hypovirus (YP_009051683), Valsa ceratosperma hypovirus 1 (YP_005476604), Sclerotinia sclerotiorum hypovirus 1 (YP_004782527), Cryphonectria hypovirus 4 (YP_138519) and Setosphaeria turcica hypovirus 1 (AZT8861) (Supplementary Figure S2). In addition to the previously mentioned conserved motifs, this viral sequence presented a DEAD-like_helicase_N superfamily (smart00487) located in the interval 2722–2880 with an E-value of 3.82 × 10−9, which contains a DEAD box helicase motif DEFH, located at position 2818–2821.
- Mymonaviridae
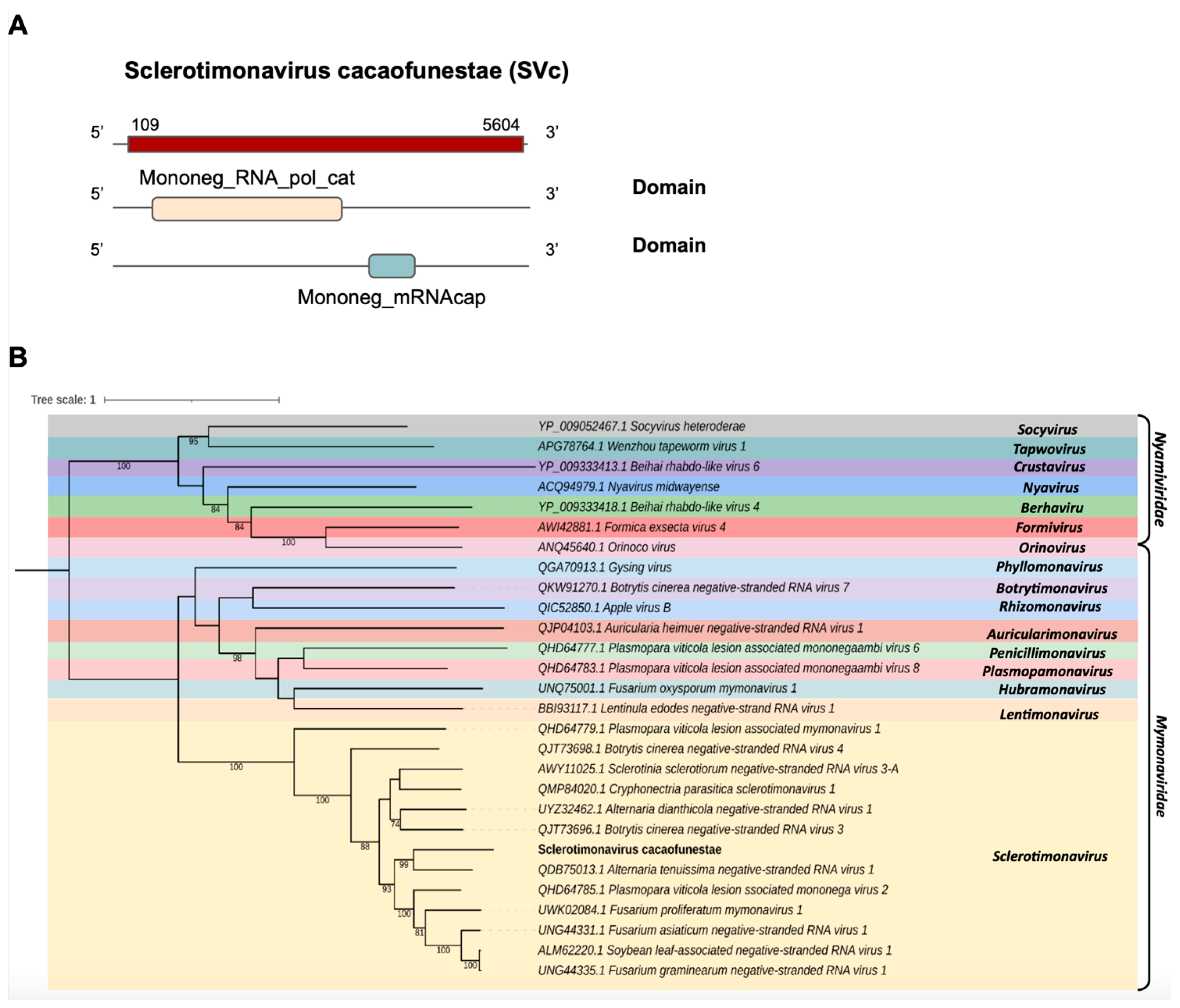
One transcript assembled from the C. cacaofunesta library, Cc_Contig2, with a size of 5.652 nt, showed similarity to viruses from the Mymonaviridae family at the amino acid level. Structural annotation indicated a large ORF of 5496 nt surrounded by 5′ and 3′ UTRs of 109 nt and 48 nt, respectively (Figure 3A). Conserved domains were identified in Cc_Contig2, such as Mononeg_RNA_pol_cat (IPR014023) and Mononeg_mRNAcap (IPR026890) (Figure 3A). Analysis of conserved domains of the closely related Alternaria tenuissima negative-stranded RNA virus 1 (AtNSRV1) revealed a very similar ORF length and the presence of the same domains identified in Cc_Contig2 (Figure 3A and Supplementary Figure S1). Phylogenetic analysis of the putative virus indicated a relationship with members of the genus Sclerotimonavirus (Mymonaviridae), closely related to Alternaria tenuissima negative-stranded RNA virus 1, with 99% bootstrap (Figure 3B). This new virus was named Sclerotimonavirus cacaofunestae (SVc).
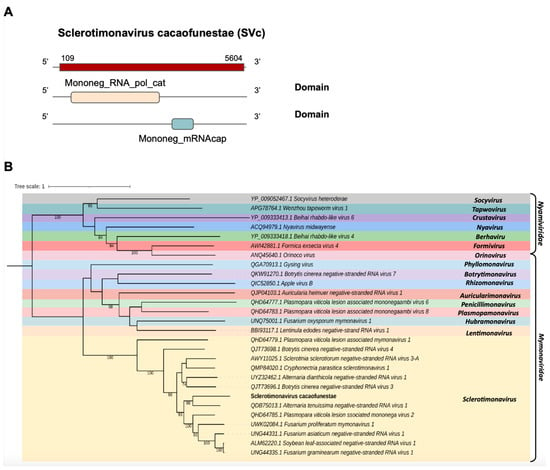
Figure 3.
Conserved domains and phylogenetic analysis of sequences showing similarity to Sclerotimonavirus. (A) Schematic representation of the novel sclerotimonavirus SVc with the ORF and domains represented. (B) Phylogenetic analysis using the ModelTest-NG according to the Akaike information criterion (AIC) indicated that the best evolutionary model was BLOSUM62 + F. Bootstrap values were established with 1000 replicates. Bootstrap values (%) lower than 70 are not shown.
- Partitiviridae
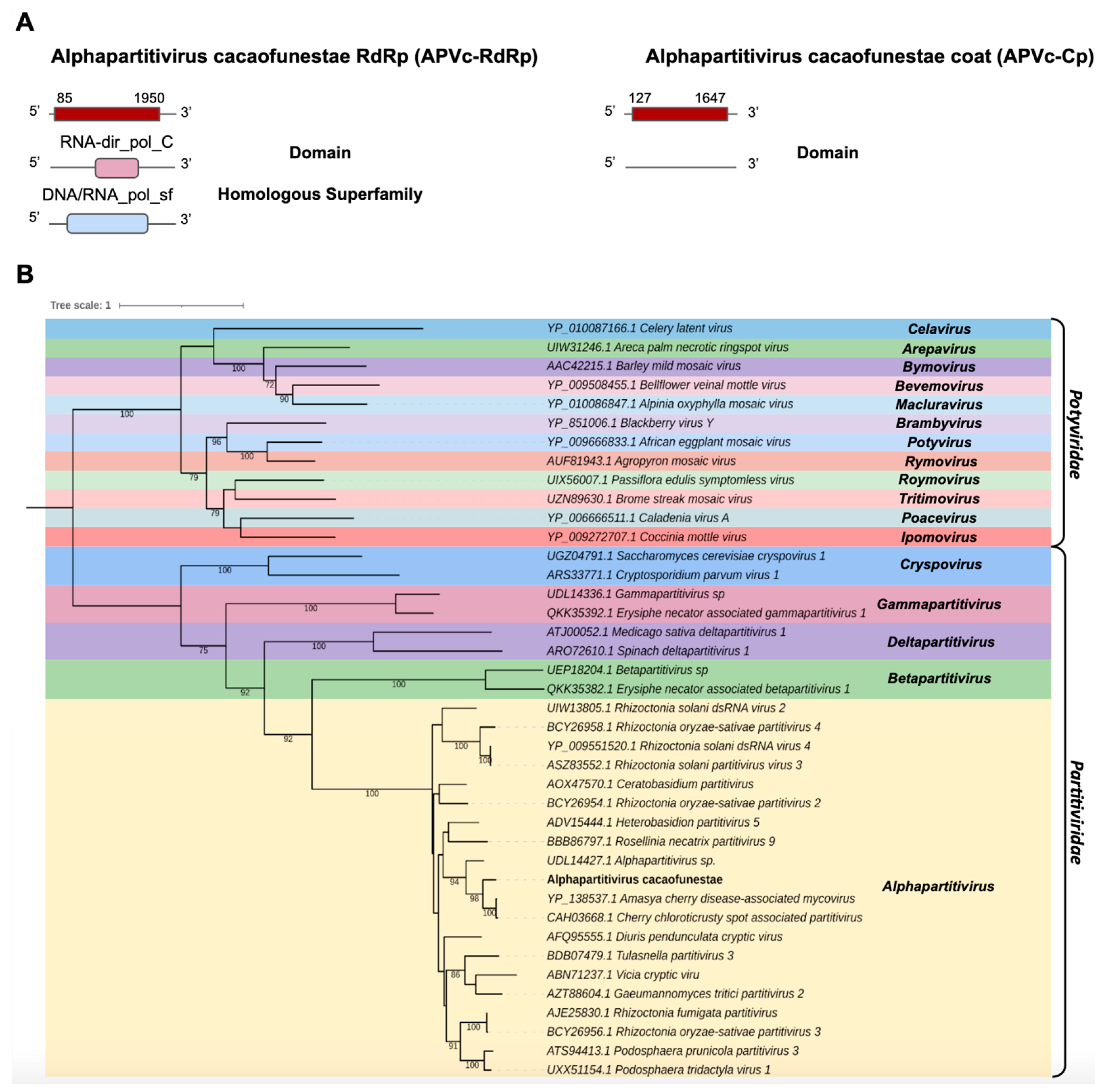
The transcript Cc_Contig3 assembled in the C. cacaofunesta library with a size of 1998 nt was closely related to viruses from the Partitiviridae family at the amino acid level. Structural annotation indicated a large ORF of 1866 nt surrounded by 5′ and 3′ UTRs of 85 nt and 48 nt, respectively. Another transcript from the C. cacaofunesta library, Cc_Contig4-Cp, with a size of 1818 nt, also showed similarity to viruses of the Partitiviridae family. This transcript was related to a coat protein whose structural annotation indicated an ORF of 1521 nt surrounded by 5′ and 3′ UTRs of 172 nt and 126 nt, respectively. Analysis of conserved domains in Cc_Contig3 revealed the presence of RNA-dir_pol_C (IPR001205) superimposed on the homologous superfamily DNA/RNA_pol_sf (IPR043502). On the other hand, Cc_Contig4-Cp did not have conserved domains (Figure 4A). Search for domains in the closest virus Amasya cherry disease-associated mycovirus (AcDAV) RdRp (currently Cherry chlorotic rusty spot associated partitivirus) revealed the presence of the same domains identified in Cc_Contig3 (Figure 4A and Supplementary Figure S1). The coat segment of AcDAV did not present domains, which was also true for Cc_contig4-Cp. According to the phylogenetic analysis, the putative virus is related to sequences from the genus Alphapartitivirus (Partitiviridae), closely related to the Amasya cherry disease-associated mycovirus (AcDAV) grouping with 98% bootstrap (Figure 4B). Therefore, the virus was named Alphapartitivirus cacaofunestae (APVc-RdRp).
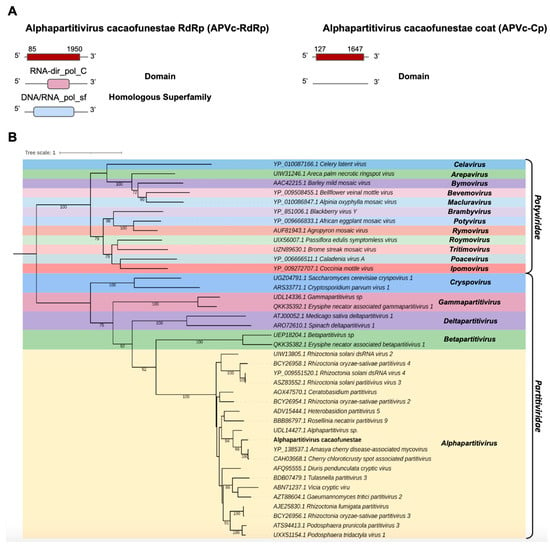
Figure 4.
Characterization of the virus related to members of the Partitiviridae family. (A) Schematic representation of the novel partitivirus segments APVc-RdRp and APVc-Cp with the ORF and domains represented. (B) Phylogenetic analysis using the ModelTest-NG according to the Akaike information criterion (AIC) indicated that the best evolutionary model was VT + F. Bootstrap values were established with 1000 replicates. Bootstrap values (%) lower than 70 are not shown.
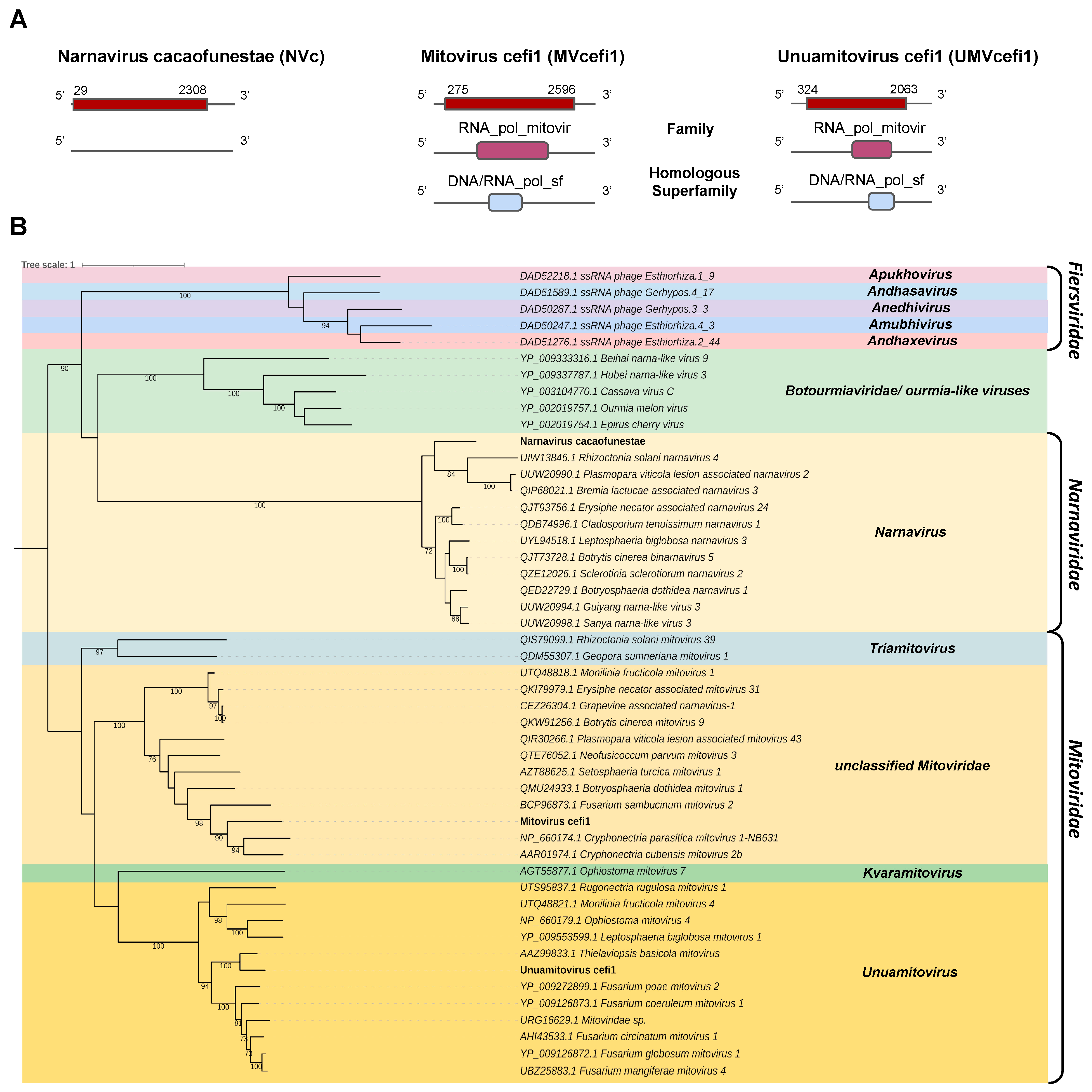
- Narnaviridae and Mitoviridae
Two transcripts assembled from the C. fimbriata library and the C. cacaofunesta library showed sequence similarity to members of the families Narnaviridae and Mitoviridae. The contig belonging to the C. cacaofunesta library, Cc_Contig5, with a size of 2409 nt, showed similarity to viruses of the Narnaviridae at the amino acid level. Structural annotation of Cc_Contig5 indicated an ORF of 2381 nt surrounded by UTRs of 3′ and 5′ with 28 nt and 102 nt, respectively. The second contig assembled from the C. fimbriata library, Cf_Contig6, presented a length of 3268 nt and showed a large ORF of 2322 nt surrounded by UTRs of 3′ and 5′ with 275 nt and 672 nt, respectively. The other contig assembled from the C. fimbriata library, Cf_Contig7, presented 2395 nt, and the structural annotation indicated an ORF of 1740 nt surrounded by UTRs of 5′ and 3′ of 324 nt and 332 nt, respectively. These two transcripts showed similarity to viruses of the Mitoviridae at the amino acid level. Of note, all three transcripts only presented large ORFs in genetic code 4 (Mold, Protozoan Mitochondrial and Coelenterate Mitochondrial and the Mycoplasma/Spiroplasma) consistent with the replication strategy of narnaviruses and mitoviruses. Cc_Contig5 did not present conserved domains. However, for Cf_Contig6 and Cf_Contig7 sequences, we observed the presence of the RNA_pol_mitovir family (IPR008686), which overlapped with the homologous DNA/RNA_pol_sf superfamily (IPR043502) (Figure 5A).
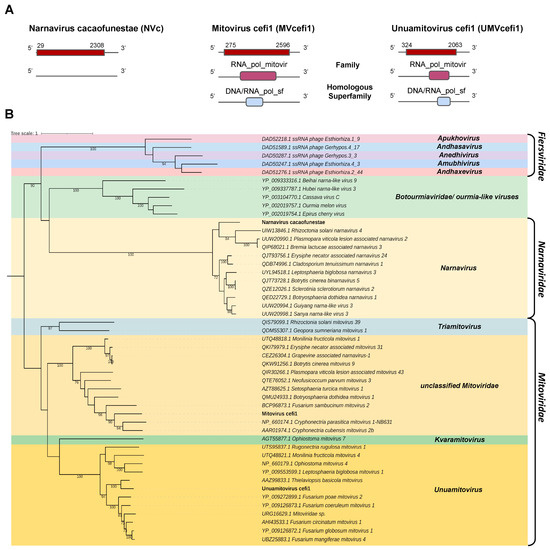
Figure 5.
Phylogenetic analysis of sequences showing similarity to Narnavirus, Mitovirus and Unuamitovirus. (A) Schematic representation of the novel narnavirus NVc and mitoviruses MVcefi1 and UMVcefi1 with the ORF and domains represented. (B) Phylogenetic analysis using the ModelTest-NG according to the Akaike information criterion (AIC) indicated that the best evolutionary model was VT + F. Bootstrap values were established with 1000 replicates. Bootstrap values (%) lower than 70 are not shown.
Analysis of conserved domains of the closely related Botrytis cinerea binarnavirus 5 (BcBNV5), Cryphonectria parasitica mitovirus 1-NB63 1(CpMV1) and Thielaviopsis basicola mitovirus (TbMV) revealed the same domain profile observed in the transcripts Cc_Contig5, Cc_Contig6 and Cc_Contig7, respectively. The exception was the RT_dom domain (IPR000477), only identified in Thielaviopsis basicola mitovirus (TbMV) (Figure 5A and Supplementary Figure S1).
According to the phylogenetic analysis, the putative virus represented by the transcript Cc_Contig5 was grouped with members of the genus Narnavirus (Narnaviridae) within the clade containing BcBNV5 (Figure 5B). Cf_Contig6 formed a clade containing members of Mitoviridae but without genus designation. Finally, Cf_Contig7 was grouped with sequences from the genus Unuamitovirus (Mitoviridae), with 100% bootstrap. (Figure 5B). These new viruses were named Narnavirus cacaofunestae (NVc), Mitovirus cefi1 (MVcefi1) and Unuamitovirus cefi1 (UMVcefi1), respectively.
Further analysis regarding conserved motifs revealed that the NVc viral transcript presented a central conserved RNA-dependent RNA polymerase (RdRp) catalytic domain ps-ssRNAv_RdRp-like superfamily (cl40470), located in the interval 527–736 with an E-value of 2.35 × 10−7. This region contained two conserved polymerase motifs called conserved polymerase A motifs, located at positions 612 and 631–644 D..FSSTDYQEATDAMQ, and conserved polymerase B motifs, located at positions 698–724 and 726–729 VLMGDPLTKPVLHLVNI..VRRI. Both motifs were found in the ps-ssRNAv_Botourmiaviridae_RdRp domain (cd23183) belonging to the Botourmiaviridae family, order Ourlivirales (Supplementary Figure S2).
- Endornaviridae
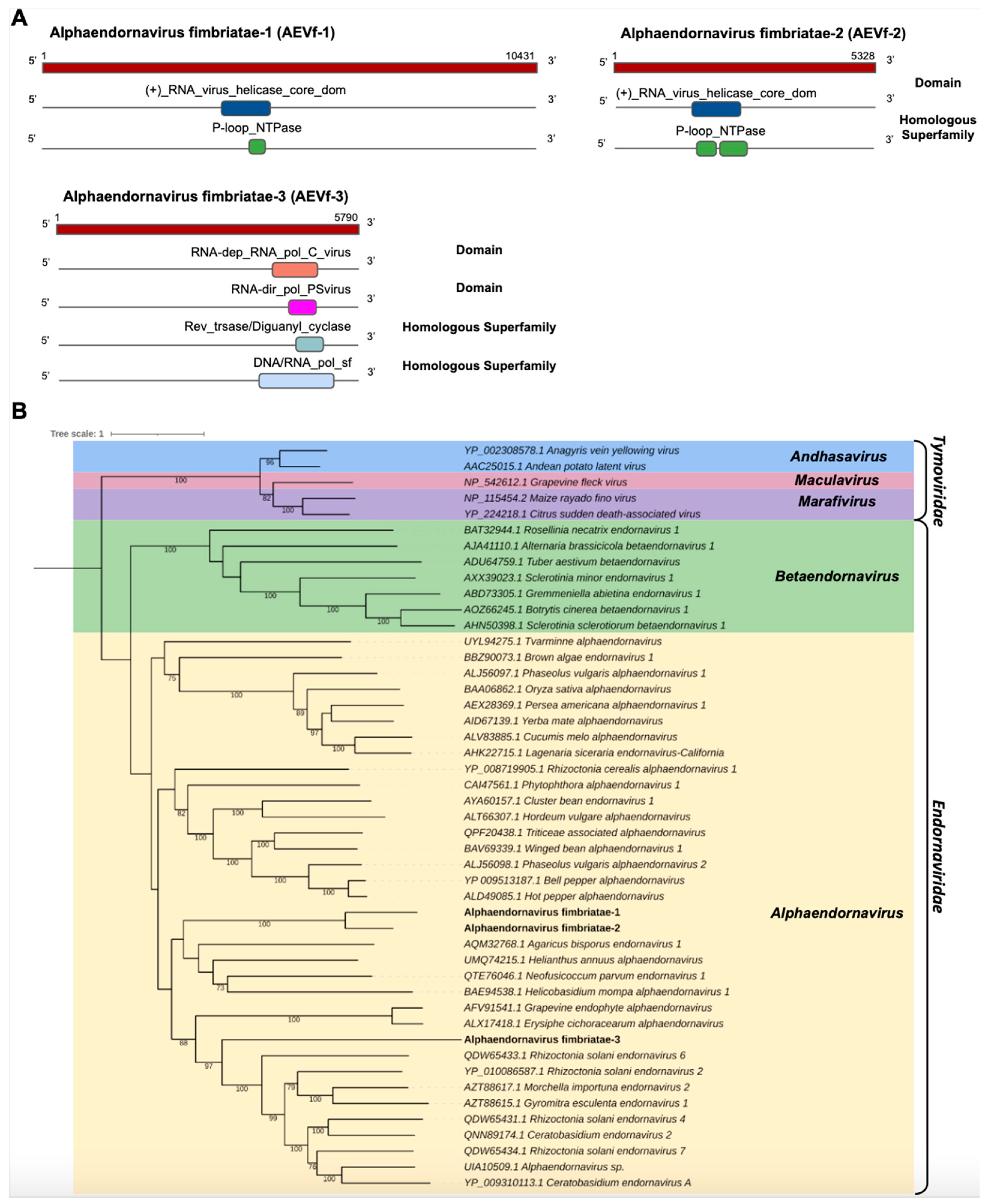
We identified three transcripts in the C. fimbriata library showing sequence similarity to members of the Endornaviridae family. Cf_Contig8, with a length of 10,432 nt, presented an ORF of 10,431 nt surrounded by UTRs of 5′ and 3′ with 1 nt and 834 nt, respectively. For Cf_Contig9, we only reconstituted a fragment with an expected length of 5328, likely representing a fragment of the large polyprotein gene. Similarly, Cf_Contig10 of 5790 nt denoted a partial ORF. All of them showed similarity to Endornaviruses at the amino acid level. Regarding the presence of conserved domains, Cf_Contig8 and Cf_Contig9 contained the RNA_virus_helicase_core_dom domain (IPR027351) superimposed on the homologous superfamily P-loop_NTPase (IPR027417), while Cf_Contig10 showed the RNA-dep_RNA_pol_C_virus (IPR001788) domain with integration of the conserved RNA-dir_pol_PSvirus (IPR007094) domains found to overlap with the homologous superfamily Rev_trsase/Diguanyl_cyclase (IPR043128) integrated with DNA/RNA_pol_sf (IPR043502) (Figure 6A).
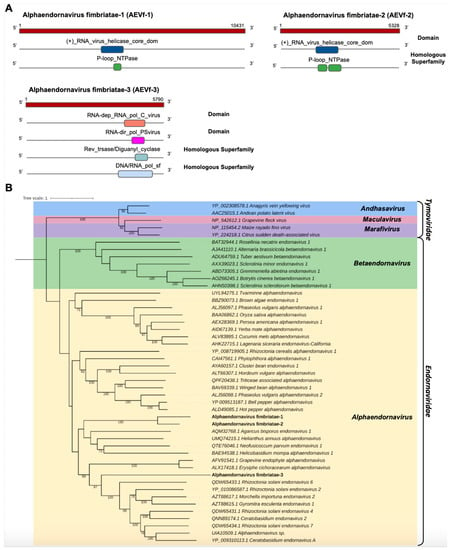
Figure 6.
Conserved domains and phylogenetic analysis of sequences showing similarity to Alphaendornavirus. (A) Schematic representation of the novel alphaendornaviruses AEVf-1, AEVf-2 and AEVf-3 with the ORF and domains represented. (B) Phylogenetic analysis using the ModelTest-NG according to the Akaike information criterion (AIC) indicated that the best evolutionary model was BLOSUM62 + F. Bootstrap values were established with 1000 replicates. Bootstrap values (%) lower than 70 are not shown.
Analysis of conserved domains of the Tvarminne alphaendornavirus, the closest viruses to Cf_Contig8, revealed the presence of shared domains, with the exception of the RNA-dep_RNA_pol_C_virus (IPR001788), RNA-dir_pol_PSvirus (IPR007094) and DNA/RNA_pol_sf (IPR043502) domains. The analysis of conserved domains of Alphaendornavirus sp., the closest virus to Cf_Contig9, revealed that they share two domains, with the exception of Mycovirus_RNAse (IPR046747), RNA-dep_RNA_pol_C_virus (IPR001788), RNA-dir_pol_PSvirus (IPR007094) and DNA/RNA_pol_sf (IPR043502). The analysis of conserved domains of Morchella importuna endornavirus 2 (MiEV2), the closest virus to Cf_Contig10, revealed that they also share two domains, with the exception of (+)_RNA_virus_helicase_core_dom (IPR027351) and P-loop_NTPase (IPR027417) only identified in MiEV2 and RNA-dir_pol_PSvirus (IPR007094) and Rev_trsase/Diguanyl_cyclase (IPR043128) only identified in Cf_Contig10 (Figure 6A and Supplementary Figure S1).
The phylogeny of the putative viruses revealed that Cf_Contig8 and Cf_Contig9 were grouped together and, like Cf_Contig10, were closely related to members of the Alphaendornavirus genus (Endornaviridae) (Figure 6B). These new viruses were named Alphaendornavirus fimbriatae-1 (AEVf-1), Alphaendornavirus fimbriatae-2 (AEVf-2) and Alphaendornavirus fimbriatae-3 (AEVf-3), respectively.
AEVf-3 also presented a conserved central catalytic domain of the RNA-dependent RNA polymerase (RdRp) ps-ssRNAv_RdRp-like super family (cl40470), located in the interval 1491–1736 with an E-value of 1.52 × 10−118. This region represents conserved polymerase motifs called conserved polymerase A motif, located at position 2613–2627 LFVEDDLEKQDRQTD; conserved polymerase B motif, located at position 1673–1696 RLTGQATTALGNVITNLLVHSRLV; and conserved polymerase C motif, located at position 1704–1718 VIMMCLGDDNLMVCR. All motifs are found in the Endornaviridae_RdRp (cd23255) domain, belonging to the Endornaviridae family (Supplementary Figure S4).
- Totiviridae
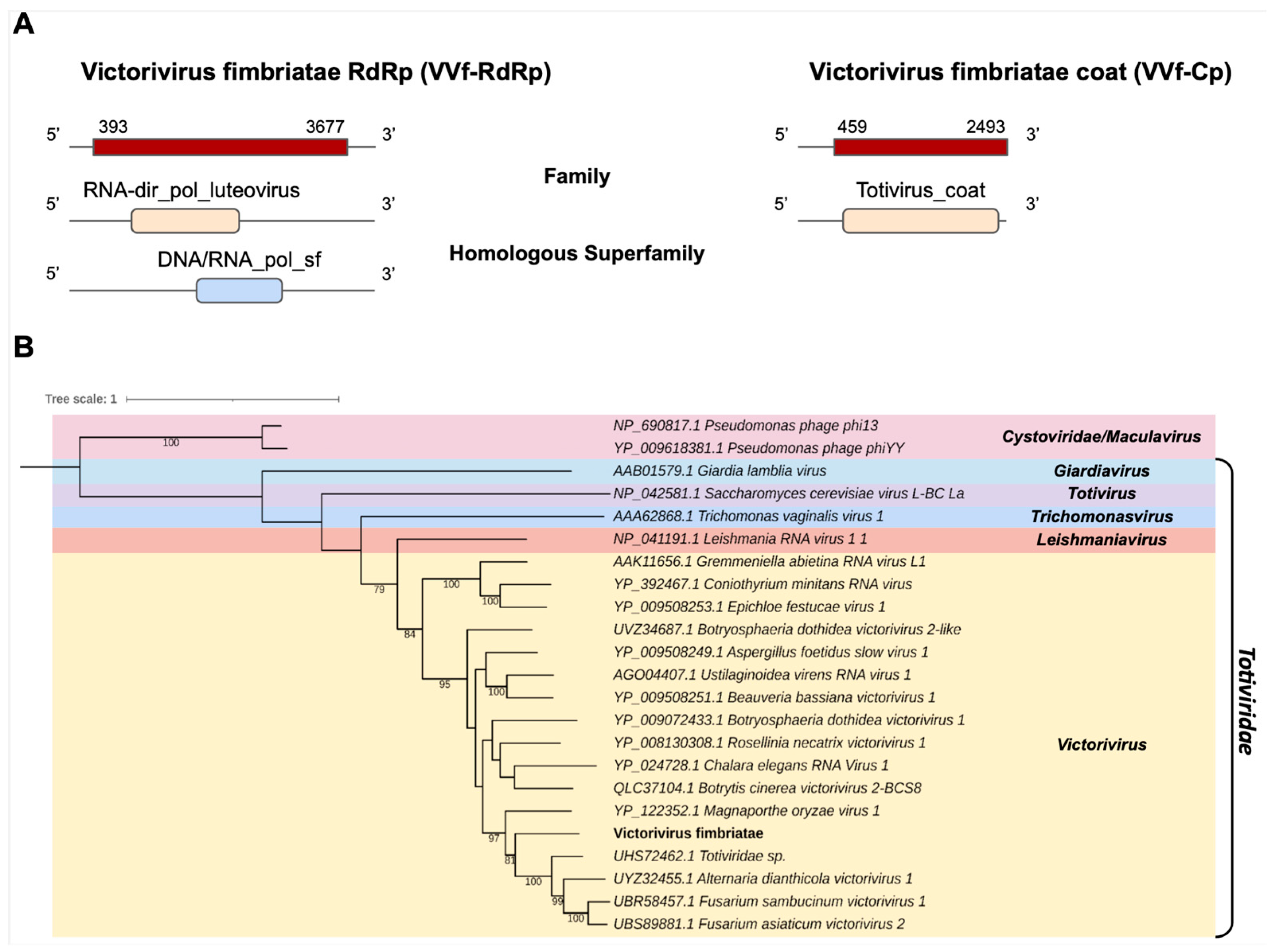
One of the transcripts assembled in the C. fimbriata library, Cf_Contig11, with a length of 3995 nt was related to viruses from the Totiviridae family at the amino acid level. Cf_Contig11 contained an ORF of 3285 nt surrounded by 5′ and 3′ UTRs with 393 nt and 318 nt, respectively. We observed that the presence of the conserved domain RNA-dir_pol_luteovirus family (IPR001795) overlapped with the homologue superfamily DNA/RNA_pol_sf (IPR043502). Another transcript in the C. fimbriata library, Cf_Contig12, with a size of 2493 nt also showed similarity to viruses of the Totiviridae at the amino acid level, with an ORF of 2034 nt surrounded by 5′ and 3′ UTRs with 459 nt and 1 nt, respectively, and presenting the conserved domain for the Totivirus_coat family (IPR008871) (Figure 7A). The analysis of conserved domains of the closely related Totiviridae sp. sequence revealed the presence of the same domains observed for Cf_Contig11 (Figure 7A and Supplementary Figure S1).
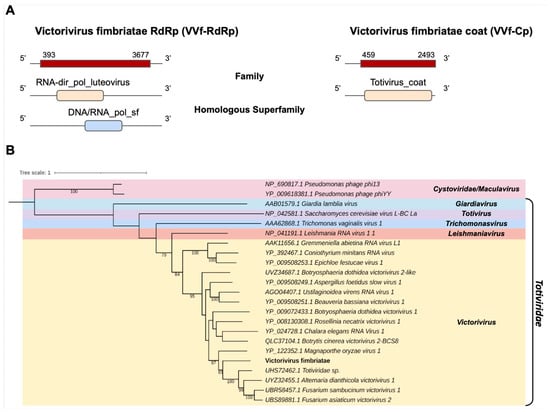
Figure 7.
Conserved domains and phylogenetic analysis of sequence showing similarity to Victorivirus. (A) Schematic representation of the novel victorivirus segments VVf-RdRp and VVf-Cp with the ORF and domains represented. (B) Phylogenetic analysis using the ModelTest-NG according to the Akaike information criterion (AIC) indicated that the best evolutionary model was BLOSUM62 + F. Bootstrap values were established with 1000 replicates. Cystovirus was used as an outgroup to root the tree. Bootstrap values (%) lower than 70 are not shown.
According to the phylogenetic analysis, the putative virus was grouped with sequences from the genus Victorivirus (Totiviridae), closely related to Totiviridae sp. (Figure 7B). This new virus was named Victorivirus fimbriatae (VVf).
3.4. Abundance of Viral Sequences
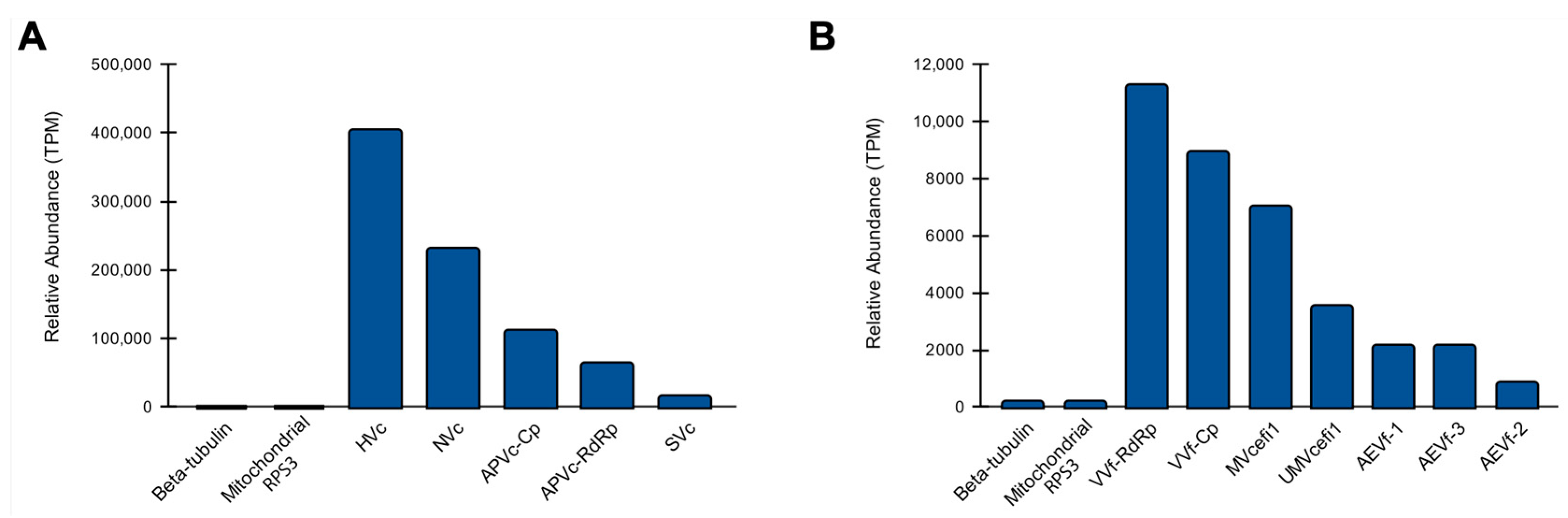
Unexpectedly, we observed a complex diversity of viruses infecting the fungal species. Therefore, we decided to investigate the abundance of these viral species in comparison with endogenous nuclear and mitochondrial genes. Regarding the putative viruses identified in C. cacaofunesta, HVc showed the highest abundance with 404.689 TPM, while SVc presented the lowest with 15.258 TPM (Figure 8A). Of note, the constitutive genes beta-tubulin and mitochondrial RPS3 accounted for 0.081 and 0.253 TPM, respectively. Our results suggest that the transcriptional activity of the C. cacaofunesta-infecting viruses is ~180 to ~5000 times higher than the constitutive nuclear gene (Figure 8A). In the case of viruses identified in C. fimbriata, VVf had the greatest abundance with 11.289 TPM, while AEVf-2 showed the lowest with 0.846 TPM (Figure 8B). Unlike what was observed for C. cacaofunesta, C. fimbriata-infecting viruses showed a transcriptional activity closer to that detected for the marker genes beta-tubulin and mitochondrial RPS3, with less than a 10-fold difference (Figure 8B).
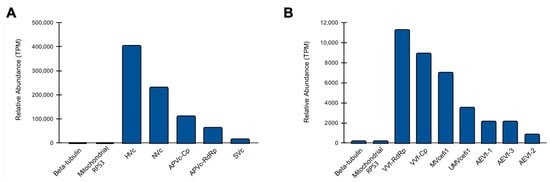
Figure 8.
Abundance of viral sequences in Ceratocystis samples. Relative abundance of new viruses identified in C. cacaofunesta (A) and C. fimbriata (B). For both organisms, we included the quantification of the constitutive genes beta-tubulin and mitochondrial RPS3.
3.5. Characterization of RNA-Interference-Related Genes in Ceratocystis Fimbriata
Other studies have shown that fungi can contain RNA interference (RNAi) machinery with important roles in genome stability and antiviral response [32]. Therefore, we hypothesized that RNAi could be involved in the management of multiple viral infections by fungi. Thus, we investigated whether their genomes contained the RNAi core genes and whether they were transcriptionally active. Since the genome of C. cacaofunesta is not annotated, we performed characterization of RNAi genes in C. fimbriata. Using proteins identified and validated experimentally in the model fungus Neurospora crassa, we searched for RNAi-related genes including the RNA-dependent RNA polymerases QDE-1 and SAD-1, Argonaute-2 orthologs, QDE-2 and SMS2 and Dicer, among others (Supplementary Table S5 and Figure S5A). The RNAi-related proteins identified had similar lengths and presence of conserved domains, including all canonical elements expected for Argonaute and Dicer genes (Supplementary Table S5 and Supplementary Figure S5A). Quantification of RNAi-related genes detected transcriptional activity for all of the genes assessed, with Argonaute orthologues showing higher abundance and SAD and SMS2 presenting the lowest number of transcripts detected.
4. Discussion
Ceratocystis in general accommodates many important pathogens of agricultural crops and woody plants. C. fimbriata was the first to be described and can be found in plantations mainly of Ipomoea batatas [11,72,73,74], Mangifera indica [75,76], Eucalyptus sp. [75,77,78], Ficus carica [75,79], Punica granatum [80,81], Colocasia esculenta [80,82,83], Theobroma cacao [11], Gmelina arborea [84,85], Syngonium sp. [86,87] and Ilex paraguariensis [88,89]. In contrast, C. cacaofunesta has only been described in Theobroma cacao [90] and in species from the genus Herrania sp. [91].
Metatranscriptomic analyses indicated the presence of several microorganisms in the libraries, including elements derived from bacteria and viruses. This diversity of microorganisms may be related to the culture medium used for fungal growth, which was enriched with cocoa xylem of C. cacaofunesta [10] and was extracted from an isolate preserved in a culture collection and subjected to genome sequencing of C. fimbriata [39,40]. Nevertheless, the possible presence of these species does not interfere with the result, since the viral sequences assembled in our study are from families described as infecting fungi, not bacteria. Furthermore, their abundance was hundreds of times lower than that of C. cacaofunesta and C. fimbriata, inconsistent with the abundance of viruses in the sample.
Hypoviridae is a family of capsidless viruses that present a positive-sense ssRNA genome of 9.1 to 12.7 kb encoding one or two ORFs [92,93]. These characteristics are similar to those found in our research. In addition, when comparing the sequence called HVc with Mycosphaerella hypovirus A, we observed similarities in terms of the conserved domains found, demonstrating the probable membership of the new virus in the Hypoviridae family. It is worth mentioning that other studies have identified Hypovirus in other fungal species, such as the brown rot fungus Monilinia fructicola reported by De Miccolis Angelini and collaborators (2022) [94], who named them Monilinia fructicola hypovirus 1 and Monilinia fructicola hypovirus 2, or MfrcHV1 and MfrcHV2, respectively. In MfrcHV1, the polyprotein had three conserved domains (an RdRp, a putative peptidase PPPDE and a C-terminal helicase), while MfrcHV2 had two conserved domains in the polyproteins (the superfamilies 1 and 2 of type 1 ATP binding and C-terminal helicases), presenting similarity with the conserved domains present in the viral contigs described in our study.
Other researchers who have reported the identification and characterization of hypoviruses are Velasco and collaborators (2018) [95], studying Entoleuca sp.; Li and collaborators (2020) [96], studying Bipolaris oryzae; Wang and collaborators (2022) [97], who studied Fusarium graminearum hypovirus 1 (FgHV1) and demonstrated that it has proteins with characteristics such as suppression of host RNA silencing; and Aulia and collaborators (2021) [98], who studied the fungus Cryphonectria parasitica and showed that it facilitates infection by inducing hypovirulence. The viral contig HVc was the first hypovirus detected in Ceratocystis cacaofunesta.
The viruses from the Mymonaviridae produce filamentous and enveloped virions containing a single negative-sense linear RNA molecule of about 10 kb. Mimonaviruses usually infect filamentous fungi, and one of the negative-strand RNAs of one virus, Sclerotinia sclerotiorum virus 1, induces hypovirulence in the fungal host [99]. Our phylogenetic analysis suggests a new virus genome belonging to this family, SVc. Furthermore, a previous study by Li and collaborators (2022) [100] characterized a new ssRNA(−) mycovirus isolated from the fungus Auricularia heimuer CCMJ1222, while another study, by Hao, Wu & Li (2018) [101], described an isolate of the phytopathogenic fungus Botrytis cinerea. The viral contig SVc was the first sclerotimonavirus detected in Ceratocystis cacaofunesta.
According to Vainio and collaborators (2018) [102], Partitiviridae is a family of small, isometric, nonenveloped viruses with bisegmented double-stranded (ds) RNA genomes of 3–4.8 kbp. The two genome segments are individually encapsulated and have a single large ORF or two ORFs, as in the current study where an ORF was presented, identifying and characterizing a new virus belonging to the Partitiviridae family, which presented a fragment of RdRp and a coat protein segment, which were named APVc-RdRp and APVc-CP, respectively, due to their phylogenetic proximity to alphapartitiviruses. Furthermore, Jiang and collaborators (2021) [103] studied the fungus Aspergillus nidulans, and Moriyama and collaborators (2021) [104] analyzed the phytopathogen Rosellinia necatrix, the causative agent of root rot, also identifying viral sequences related to this family. Finally, Deng & Boland (2007) [105] reported viruses belonging to the genus Partiviridae isolated from Ceratocystis resinifera, called named Ceratocystis partitivirus 1 CPV1. This virus has the same characteristics as the virus present in Ceratocystis polonica. The viral contigs APVc-RdRp and APVc-CP represent the first alphapartitivirus detected in Ceratocystis cacaofunesta.
In addition, a new viral contig called NVc belonging to Narnaviridae was identified. The contig showed similarity to individual RNA virus genomes ranging from 2.3 to 3.6 kb encoding only a single polypeptide having an RdRp domain. This feature was found in the study by Botella and collaborators (2022) [106], who described narnaviruses in the fungus Phytophthora palustris, viruses that are related to the traditional positive-coding monopartite, included in the genus Narnavirus. Also of note, Kinsella and collaborators (2022) [107] identified narnaviruses in the hosts R. oryzae and A. lentulus, which are pathogenic fungi, and Zou et al. (2021) [108] analyzed the fungus Botryosphaeria dothidea, which is the causal agent of the disease pear ring rot. The viral contig NVc is the first narnavirus detected in Ceratocystis cacaofunesta.
Two new viral contigs, named UMVcefi1 and MVcefi1, belonging to the Mitoviridae were characterized in our study. Members of this family are positive-stranded ssRNA viruses of 2.5 to 2.9 kb. Generally, the genome has a single ORF encoding an RdRp. Previous studies have also identified viral contigs in different fungal phytopathogens, such as Wang and collaborators (2021 A-B) [109,110] and de Rezende and collaborators (2021) [111]. Shafik and collaborators (2021) [112] identified a new mycovirus, named Melanconiella theae mitovirus 1 (MtMV1), a virus that infects the phytopathogenic MtMV1 and shares most of the characteristics of the members of the Mitoviridae family. The UMVcefi1 and MVcefi1 sequences represents the first mitoviruses detected in Ceratocystis fimbriata.
AEVf-1, AEVf-2 and AEVf-3 are similar to Endornaviridae, which has a linear dsRNA genome of about 14 kb to 17.6 kb. This genome encodes an ORF potentially divided into several polypeptides. Here, two major ORF-associated domains were found: Tymovirus, RdRp, and (+) RNA virus helicase core dom, which have been identified in several virus types [113] including a variety of plant virus families [114]. In addition, Luo and collaborators (2022) [115], analyzing S. sclerotiorum, identified a new endornavirus called Scelrotinia sclerotiorum endornavirus 11 (SsEV11), suggesting that there is remarkable phylogenetic diversity in the Endornaviridae family and corroborating previous findings that viruses of this family can be found in fungi. In this respect, Cao and collaborators (2022) [116] studied Ceratobasidium infected with three endornaviruses with significantly upregulated micro-RNA-like RNAs (Cer-milRNAs) and identified transcription factors, suggesting an effect of viral infection on the control of regulatory networks and growth and development of the fungus. Moreover, Marais (2021) [117] investigated Neofusicoccum parvum and found the presence of co-infecting viruses from Totiviridae, Victorivirus, Endornaviridae, Mitoviridae and Narnaviridae. The viral contigs AEVf-1, AEVf-2 and AEVf-3 are the first alphaendornaviruses belonging to the Endornaviridae family detected in Ceratocystis fimbriata.
Finally, the Totiviridae has the main feature of being a nonenveloped icosahedral virion composed of a single coat protein (Cp), with a diameter of about 40 nm and a linear dsRNA genome of 4.6 to 6.7 kb. In addition, it has two overlapping ORFs, gag and pol, respectively encoding Cp and RdRp. In the viral contig assembled in our study (VVf), we identified a conserved domain of RdRp Luteovirus, which is an essential protein encoded in the genomes of all RNA containing viruses with no DNA stage [118,119]. The viral contigs VVf-RdRp and VVf-Cp were the first representants of a Victorivirus detected in Ceratocystis fimbriata.
Many fungal species have been found able to counteract multiple viral infections relying on the RNA interference (RNAi) mechanisms with antiviral activity, such as described for Cryphonectria parasitica [29]. When the host’s RNAi mechanism targets the viral genome, an overlapping population of virus-derived vsiRNA can be produced. In C. parasitica, the dicer-like (dcl-2) and argonaute-like (agl-2) genes are necessary for antiviral silencing [27,120]. Shinji Honda (2020) [121] demonstrated that in the fungus Neurospora crassa, viral infection positively regulates the transcription of the various components of RNAi, where the Dicer (DCL-1 and DCL-2) and Argonaute (QDE-2) proteins participate in the suppression of viral replication. Mekhala Maiti (2007) [122] reported that the Argonaute QDE-2 homologue was necessary for the generation of single-stranded siRNA and gene silencing in vivo.
Although various RNAi pathways have been lost in several fungal species and lineages, most species possess this ancient mechanism [27,36]. However, there are several filamentous fungi that seem to lack the necessary components for RNA silencing to occur [27], which means that by lacking this machinery to defend against mycoviruses, these viruses produce different hypovirulence factors in their hosts. Mycovirus-mediated hypovirulence is an excellent example of multitrophic interactions, making these viruses promising for the biological control of fungal diseases [26,123].
In our study, the highest abundance of RNAi occurred in two Argonaute genes, QDE2 and SMS2, with a value of around 300 TPM, as previously indicated by Lee and collaborators (2009) [124] in their study of QDE genes induced by DNA damage. On the other hand, Li and collaborators. (2022) [100], studying Fusarium oxysporum f. sp. cubense, noted that by knocking out QDE2, the fungus showed reduced virulence, suggesting its involvement in pathogenesis. Another noteworthy fact is that the biogenesis of milR4 requires MRPL3 [32], which may explain its presence in the data analyzed. We can conclude that the genes have all the components of the RNAi pathway and are transcriptionally active, suggesting that the pathway may help the fungus combat viral infection.
Overall, of the Ceratocystis species reported to date, two stand out: Ceratocystis fimbriata, which has been reported to infect nearly 100 host species, including many woody plants [125,126], and Ceratocystis cacaofunesta, which has been reported in Theobroma cacao and in the genus Herrania [90,125,126]. Therefore, our study reveals new insights into the mycovirome diversity of Ceratocystis, an important pathogen that causes irreversible damage to the vascular system of cocoa plants, reducing yield and, in severe cases, killing the plant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12020287/s1, Table S1: Overview of RNA deep sequencing libraries used in the work; Table S2: Best hits at nucleotide and amino acid levels for viral transcripts assembled in our work; Table S3: Viral genomes used in phylogenetic analyses; Table S4: Metagenomic analysis of RNA sequencing libraries using Kaiju software; Table S5: RNAi-related genes; Figure S1: Genomic characteristics of viral genomes closely related to the viruses identified in our work; Figure S2: Conserved motifs and catalytic site in the Hypovirus cacaofunestae viral sequence; Figure S3: Conserved motifs and catalytic site in the Narnavirus cacaofunestae viral sequence; Figure S4: Conserved motifs and catalytic site in the Alphaendornavirus fimbriatae-3 viral sequence; Figure S5: Characterization of RNAi-related genes in Ceratocystis fimbriata.
Author Contributions
Conceptualization, E.R.G.R.A.; methodology, R.B.A.E., S.F.d.S., P.L.C.F., R.J.S.S. and V.C.S.; software, R.B.A.E., S.F.d.S. and V.C.S.; validation, R.B.A.E., S.F.d.S., P.L.C.F. and E.R.G.R.A.; formal analysis, R.B.A.E., S.F.d.S., P.L.C.F. and E.R.G.R.A.; investigation, R.B.A.E., S.F.d.S. and P.L.C.F.; resources, C.P.P. and E.R.G.R.A.; data curation, R.B.A.E. and S.F.d.S.; writing—original draft preparation, R.B.A.E., S.F.d.S., G.N.R.L., R.X.C., L.L.L., C.P.P., A.G.-N. and E.R.G.R.A.; writing—review and editing, R.B.A.E., S.F.d.S. and E.R.G.R.A.; visualization, R.B.A.E. and S.F.d.S.; supervision, E.R.G.R.A.; project administration, E.R.G.R.A.; funding acquisition, E.R.G.R.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated and analyzed during the current study are available in the NCBI GenBank Third Party Annotation database under accession numbers BK062942-BK062946 and BK063053-BK063059.
Acknowledgments
We thank the all the members of Virus Bioinformatics Laboratory–UESC for the fruitful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bekele, F.; Phillips-Mora, W. Cacao (Theobroma Cacao L.) Breeding. In Advances in Plant Breeding Strategies: Industrial and Food Crops: Volume 6; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 409–487. [Google Scholar] [CrossRef]
- Fernandes, L.D.S.; Royaert, S.; Corrêa, F.M.; Mustiga, G.M.; Marelli, J.-P.; Corrêa, R.X.; Motamayor, J.C. Mapping of a Major QTL for Ceratocystis Wilt Disease in an F1 Population of Theobroma Cacao. Front. Plant Sci. 2018, 9, 155. [Google Scholar] [CrossRef] [PubMed]
- Marelli, J.-P.; Guest, D.I.; Bailey, B.A.; Evans, H.C.; Brown, J.K.; Junaid, M.; Barreto, R.W.; Lisboa, D.O.; Puig, A.S. Chocolate Under Threat from Old and New Cacao Diseases. Phytopathology 2019, 109, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- International Cocoa Organization—Statistics 2021. Available online: https://www.icco.org/statistics/ (accessed on 17 October 2021).
- Hussain, B. Modernization in Plant Breeding Approaches for Improving Biotic Stress Resistance in Crop Plants. Turk. J. Agric. For. 2015, 39, 515–530. [Google Scholar] [CrossRef]
- Ploetz, R. The Impact of Diseases on Cacao Production: A Global Overview. In Cacao Diseases: A History of Old Enemies and New Encounters; Bailey, B.A., Meinhardt, L.W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 33–59. [Google Scholar] [CrossRef]
- Cabrera, O.G.; Molano, E.P.L.; José, J.; Álvarez, J.C.; Pereira, G.A.G. Ceratocystis Wilt Pathogens: History and Biology—Highlighting C. Cacaofunesta, the Causal Agent of Wilt Disease of Cacao. In Cacao Diseases: A History of Old Enemies and New Encounters; Bailey, B.A., Meinhardt, L.W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 383–428. [Google Scholar] [CrossRef]
- Delgado-Ospina, J.; Molina-Hernández, J.B.; Chaves-López, C.; Romanazzi, G.; Paparella, A. The Role of Fungi in the Cocoa Production Chain and the Challenge of Climate Change. J. Fungi 2021, 7, 202. [Google Scholar] [CrossRef]
- Santos, S.A.; Vidigal, P.M.P.; Thrimawithana, A.; Betancourth, B.M.L.; Guimarães, L.M.S.; Templeton, M.D.; Alfenas, A.C. Comparative Genomic and Transcriptomic Analyses Reveal Different Pathogenicity-Related Genes among Three Eucalyptus Fungal Pathogens. Fungal Genet. Biol. 2020, 137, 103332. [Google Scholar] [CrossRef]
- Molano, E.P.L.; Cabrera, O.G.; Jose, J.; do Nascimento, L.C.; Carazzolle, M.F.; Teixeira, P.J.P.L.; Alvarez, J.C.; Tiburcio, R.A.; Tokimatu Filho, P.M.; de Lima, G.M.A.; et al. Ceratocystis Cacaofunesta Genome Analysis Reveals a Large Expansion of Extracellular Phosphatidylinositol-Specific Phospholipase-C Genes (PI-PLC). BMC Genom. 2018, 19, 58. [Google Scholar] [CrossRef]
- Engelbrecht, C.J.B.; Harrington, T.C. Intersterility, Morphology and Taxonomy of Ceratocystis Fimbriata on Sweet Potato, Cacao and Sycamore. Mycologia 2005, 97, 57–69. [Google Scholar] [CrossRef]
- Raffa, K.F.; Hobson, K.R.; Lafontaine, S.; Aukema, B.H. Can Chemical Communication Be Cryptic? Adaptations by Herbivores to Natural Enemies Exploiting Prey Semiochemistry. Oecologia 2007, 153, 1009–1019. [Google Scholar] [CrossRef]
- Smith, S.M.; Hulcr, J. Chapter 12—Scolytus and Other Economically Important Bark and Ambrosia Beetles. In Bark Beetles; Vega, F.E., Hofstetter, R.W., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 495–531. [Google Scholar] [CrossRef]
- da Cruz, M.B.; de Sousa, D.F.; de Almeida Oliveira, L.; de França, J.P.; de França, L.P.; Conceição, A.O.; Fonseca Chaves, A.L. Interference of Aqueous and Ethanolic Solutions of Adiantum Latifolium Lam. (Pteridaceae) Leaves on in Vitro Ceratocystis Cacaofunesta Mycelial Growth. Arq. Inst. Biol. 2019, 86. [Google Scholar] [CrossRef]
- Mora-Ocampo, I.Y.; Pirovani, C.P.; Luz, E.D.M.N.; Rêgo, A.P.B.; Silva, E.M.A.; Rhodes-Valbuena, M.; Corrêa, R.X. Ceratocystis Cacaofunesta Differentially Modulates the Proteome in Xylem-Enriched Tissue of Cocoa Genotypes with Contrasting Resistance to Ceratocystis Wilt. Planta 2021, 254, 94. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, T.; Xu, M.; Guo, J.; Zhang, C.; Feng, Z.; Peng, X.; Li, Z.; Xing, K.; Qin, S. Antifungal Effect of Volatile Organic Compounds Produced by Pseudomonas Chlororaphis Subsp. Aureofaciens SPS-41 on Oxidative Stress and Mitochondrial Dysfunction of Ceratocystis Fimbriata. Pestic. Biochem. Physiol. 2021, 173, 104777. [Google Scholar] [CrossRef] [PubMed]
- Aime, M.C.; Phillips-Mora, W. The Causal Agents of Witches’ Broom and Frosty Pod Rot of Cacao (Chocolate, Theobroma Cacao) Form a New Lineage of Marasmiaceae. Mycol. 2005, 97, 1012–1022. [Google Scholar] [CrossRef]
- Surujdeo-Maharaj, S.; Sreenivasan, T.N.; Motilal, L.A.; Umaharan, P. Black Pod and Other Phytophthora Induced Diseases of Cacao: History, Biology, and Control. In Cacao Diseases: A History of Old Enemies and New Encounters; Bailey, B.A., Meinhardt, L.W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 213–266. [Google Scholar] [CrossRef]
- Rodrigues, G.d.S.; Magalhães, D.M.A.; da Costa, A.M.; Luz, E.D.M.N. Antagonism of Trichoderma Spp. To the Etiological Agent of Ceratocystis Wilt in Cacao. Summa Phytopathol. 2018, 44, 72–78. [Google Scholar] [CrossRef]
- Torres-Trenas, A.; Prieto, P.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Mycovirus Fusarium Oxysporum f. Sp. Dianthi Virus 1 Decreases the Colonizing Efficiency of Its Fungal Host. Front Cell Infect. Microbiol. 2019, 9, 51. [Google Scholar] [CrossRef]
- Sharma, M.; Guleria, S.; Singh, K.; Chauhan, A.; Kulshrestha, S. Mycovirus Associated Hypovirulence, a Potential Method for Biological Control of Fusarium Species. Virus Dis. 2018, 29, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Wagemans, J.; Holtappels, D.; Vainio, E.; Rabiey, M.; Marzachì, C.; Herrero, S.; Ravanbakhsh, M.; Tebbe, C.C.; Ogliastro, M.; Ayllón, M.A.; et al. Going Viral: Virus-Based Biological Control Agents for Plant Protection. Annu. Rev. Phytopathol. 2022, 60, 21–42. [Google Scholar] [CrossRef]
- Wen, C.; Wan, X.; Zhang, Y.; Du, H.; Wei, C.; Zhong, R.; Zhang, H.; Shi, Y.; Xie, J.; Fu, Y.; et al. Molecular Characterization of the First Alternavirus Identified in Fusarium Oxysporum. Viruses 2021, 13, 2026. [Google Scholar] [CrossRef]
- Xiao, X.; Cheng, J.; Tang, J.; Fu, Y.; Jiang, D.; Baker, T.S.; Ghabrial, S.A.; Xie, J. A Novel Partitivirus That Confers Hypovirulence on Plant Pathogenic Fungi. J. Virol. 2014, 88, 10120–10133. [Google Scholar] [CrossRef]
- Zhu, J.Z.; Guo, J.; Hu, Z.; Zhang, X.T.; Li, X.G.; Zhong, J. A Novel Partitivirus That Confer Hypovirulence to the Plant Pathogenic Fungus Colletotrichum Liriopes. Front Microbiol 2021, 12, 653809. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Z.; Zhu, H.J.; Gao, B.D.; Zhou, Q.; Zhong, J. Diverse, Novel Mycoviruses From the Virome of a Hypovirulent Sclerotium Rolfsii Strain. Front Plant Sci. 2018, 9, 1738. [Google Scholar] [CrossRef]
- Segers, G.C.; Zhang, X.; Deng, F.; Sun, Q.; Nuss, D.L. Evidence That RNA Silencing Functions as an Antiviral Defense Mechanism in Fungi. Proc. Natl. Acad. Sci. USA 2007, 104, 12902–12906. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Lee, K.-M.; Cho, W.K.; Park, J.Y.; Kim, K.-H. Differential Contribution of RNA Interference Components in Response to Distinct Fusarium Graminearum Virus Infections. J. Virol. 2018, 92, e01756-e17. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Mycoviruses, RNA Silencing, and Viral RNA Recombination. Adv. Virus Res. 2011, 80, 25–48. [Google Scholar] [CrossRef] [PubMed]
- Nicolás, F.E.; Ruiz-Vázquez, R.M. Functional Diversity of RNAi-Associated SRNAs in Fungi. Int. J. Mol. Sci. 2013, 14, 15348–15360. [Google Scholar] [CrossRef]
- Nicolás, F.E.; Vila, A.; Moxon, S.; Cascales, M.D.; Torres-Martínez, S.; Ruiz-Vázquez, R.M.; Garre, V. The RNAi Machinery Controls Distinct Responses to Environmental Signals in the Basal Fungus Mucor Circinelloides. BMC Genom. 2015, 16, 237. [Google Scholar] [CrossRef]
- Dang, Y.; Yang, Q.; Xue, Z.; Liu, Y. RNA Interference in Fungi: Pathways, Functions, and Applications. Eukaryot. Cell 2011, 10, 1148–1155. [Google Scholar] [CrossRef]
- Liu, Y. RNA Interference Pathways in Filamentous Fungi. Cell Mol. Life Sci. 2010, 67, 3849–3863. [Google Scholar] [CrossRef]
- Khatri, M.; Rajam, M.V. Targeting Polyamines of Aspergillus Nidulans by SiRNA Specific to Fungal Ornithine Decarboxylase Gene. Med. Mycol. 2007, 45, 211–220. [Google Scholar] [CrossRef]
- Mukherjee, K.; Campos, H.; Kolaczkowski, B. Evolution of Animal and Plant Dicers: Early Parallel Duplications and Recurrent Adaptation of Antiviral RNA Binding in Plants. Mol. Biol. Evol. 2013, 30, 627–641. [Google Scholar] [CrossRef]
- Nakayashiki, H.; Kadotani, N.; Mayama, S. Evolution and Diversification of RNA Silencing Proteins in Fungi. J. Mol. Evol. 2006, 63, 127–135. [Google Scholar] [CrossRef]
- Roossinck, M.J. Metagenomics of Plant and Fungal Viruses Reveals an Abundance of Persistent Lifestyles. Front Microbiol. 2015, 5, 767. [Google Scholar] [CrossRef] [PubMed]
- Mokili, J.L.; Rohwer, F.; Dutilh, B.E. Metagenomics and Future Perspectives in Virus Discovery. Curr. Opin. Virol. 2012, 2, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Simpson, M.C.; Wilken, P.M.; Coetzee, M.P.A.; Wingfield, M.J.; Wingfield, B.D. Analysis of Microsatellite Markers in the Genome of the Plant Pathogen Ceratocystis Fimbriata. Fungal. Biol. 2013, 117, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; de Beer, Z.W.; Wingfield, B.D. IMA Genome-F 1: Ceratocystis Fimbriata: Draft Nuclear Genome Sequence for the Plant Pathogen, Ceratocystis Fimbriata. IMA Fungus 2013, 4, 357–358. [Google Scholar] [CrossRef]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Res 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- BI Ageing Clock Team; Stubbs, T.M.; Bonder, M.J.; Stark, A.-K.; Krueger, F.; von Meyenn, F.; Stegle, O.; Reik, W. Multi-Tissue DNA Methylation Age Predictor in Mouse. Genome Biol. 2017, 18, 68. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Prjibelski, A.D.; Vasilinetc, I.; Bankevich, A.; Gurevich, A.; Krivosheeva, T.; Nurk, S.; Pham, S.; Korobeynikov, A.; Lapidus, A.; Pevzner, P.A. ExSPAnder: A Universal Repeat Resolver for DNA Fragment Assembly. Bioinformatics 2014, 30, i293–i301. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Kyran, A. Fasta Statistics: Display Summary Statistics for a Fasta File 2021. Available online: https://github.com/galaxyproject/tools-iuc (accessed on 5 November 2022).
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Rombel, I.T.; Sykes, K.F.; Rayner, S.; Johnston, S.A. ORF-FINDER: A Vector for High-Throughput Gene Identification. Gene 2002, 282, 33–41. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39 (Suppl. S2), W29–W37. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER Web Server: 2015 Update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Almeida, A.; Beracochea, M.; Boland, M.; Burgin, J.; Cochrane, G.; Crusoe, M.R.; Kale, V.; Potter, S.C.; Richardson, L.J.; et al. MGnify: The Microbiome Analysis Resource in 2020. Nucleic Acids Res. 2020, 48, D570–D578. [Google Scholar] [CrossRef]
- Schulz, M.H.; Zerbino, D.R.; Vingron, M.; Birney, E. Oases: Robust de Novo RNA-Seq Assembly across the Dynamic Range of Expression Levels. Bioinformatics 2012, 28, 1086–1092. [Google Scholar] [CrossRef] [PubMed]
- Afiahayati; Sato, K.; Sakakibara, Y. MetaVelvet-SL: An Extension of the Velvet Assembler to a de Novo Metagenomic Assembler Utilizing Supervised Learning. DNA Res. 2015, 22, 69–77. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de Novo Assembler for Single-Cell and Metagenomic Sequencing Data with Highly Uneven Depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and Sensitive Taxonomic Classification for Metagenomics with Kaiju. Nat. Commun 2016, 7, 11257. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, Unit 2.3. [Google Scholar] [CrossRef]
- Han, M.V.; Zmasek, C.M. PhyloXML: XML for Evolutionary Biology and Comparative Genomics. BMC Bioinform. 2009, 10, 356. [Google Scholar] [CrossRef] [PubMed]
- Marincowitz, S.; Barnes, I.; de Beer, Z.W.; Wingfield, M.J. Epitypification of Ceratocystis Fimbriata. Fungal Syst. Evol. 2020, 6, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, M.; Wang, Y.; Li, L.; Wang, M.; Li, X.; Xu, M.; Loake, G.J.; Guo, M.; Jiang, J. Ceratocystis Fimbriata Employs a Unique Infection Strategy Targeting Peltate Glandular Trichomes of Sweetpotato (Ipomoea Batatas) Plants. Phytopathology 2020, 110, 1923–1933. [Google Scholar] [CrossRef]
- Stahr, M.; Quesada-Ocampo, L.M. Assessing the Role of Temperature, Inoculum Density, and Wounding on Disease Progression of the Fungal Pathogen Ceratocystis Fimbriata Causing Black Rot in Sweetpotato. Plant Dis. 2020, 104, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Harrington, T.C.; Thorpe, D.J.; Alfenas, A.C. Genetic Variation and Variation in Aggressiveness to Native and Exotic Hosts among Brazilian Populations of Ceratocystis Fimbriata. Phytopathology 2011, 101, 555–566. [Google Scholar] [CrossRef]
- Kumari, N.; Shukla, P.K.; Singh, H.; Fatima, T.; Bajpai, A. Development of Species-Specific PCR Based Detection Assay for Ceratocystis Fimbriata, Mango Wilt Pathogen. Indian Phytopathol. 2022, 75, 215–220. [Google Scholar] [CrossRef]
- Fourie, A.; Wingfield, M.J.; Wingfield, B.D.; Barnes, I. Molecular Markers Delimit Cryptic Species in Ceratocystis Sensu Stricto. Mycol. Prog. 2014, 14, 1020. [Google Scholar] [CrossRef]
- Harrington, T.C.; Kazmi, M.R.; Al-Sadi, A.M.; Ismail, S.I. Intraspecific and Intragenomic Variability of ITS RDNA Sequences Reveals Taxonomic Problems in Ceratocystis Fimbriata Sensu Stricto. Mycologia 2014, 106, 224–242. [Google Scholar] [CrossRef] [PubMed]
- Yakushiji, H.; Morita, T.; Jikumaru, S.; Ikegami, H.; Azuma, A.; Koshita, Y. Interspecific Hybridization of Fig (Ficus Carica L.) and Ficus Erecta Thunb., a Source of Ceratocystis Canker Resistance. Euphytica 2012, 183, 39–47. [Google Scholar] [CrossRef]
- Li, Q.; Harrington, T.C.; McNew, D.; Li, J.; Huang, Q.; Somasekhara, Y.M.; Alfenas, A.C. Genetic Bottlenecks for Two Populations of Ceratocystis Fimbriata on Sweet Potato and Pomegranate in China. Plant Dis. 2016, 100, 2266–2274. [Google Scholar] [CrossRef]
- Huang, Q.; Zhu, Y.Y.; Chen, H.R.; Wang, Y.Y.; Liu, Y.L.; Lu, W.J.; Ruan, X.Y. First Report of Pomegranate Wilt Caused by Ceratocystis Fimbriata in Yunnan, China. Plant Dis. 2003, 87, 1150. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wang, Y.Y.; Zhao, Y.Y.; Jiao, Y.X.; Li, X.F.; Chen, H.R.; Zhu, Y.Y. First Report of Taro Black Rot Caused by Ceratocystis Fimbriata in China. Plant Pathol. 2008, 57, 780. [Google Scholar] [CrossRef]
- Harrington, T.C.; Thorpe, D.J.; Marinho, V.L.A.; Furtado, E.L. First Report of Black Rot of Colocasia Esculenta Caused by Ceratocystis Fimbriata in Brazil. Fitopatol. Bras. 2005, 30, 88–89. [Google Scholar] [CrossRef]
- Baker, C.J.; Harrington, T.C.; Krauss, U.; Alfenas, A.C. Genetic Variability and Host Specialization in the Latin American Clade of Ceratocystis Fimbriata. Phytopathology 2003, 93, 1274–1284. [Google Scholar] [CrossRef]
- Méndez-Álvarez, D.; de Souza Cândido, T.; Couto Alfenas, A.; Murillo, O.; Badilla, Y.; Ferreira Alfenas, R. First Report of Ceratocystis Fimbriata Causing Wilt on Gmelina Arborea in Costa Rica. For. Pathol. 2020, 50, e12628. [Google Scholar] [CrossRef]
- Thorpe, D.J.; Harrington, T.C.; Uchida, J.Y. Pathogenicity, Internal Transcribed Spacer-RDNA Variation, and Human Dispersal of Ceratocystis Fimbriata on the Family Araceae. Phytopathology 2005, 95, 316–323. [Google Scholar] [CrossRef]
- Walker, J.; Tesoriero, L.; Pascoe, I.; Forsberg, L.I. Basal Rot of Syngonium Cultivars and the First Record of Ceratocystis Fimbriata from Australia. Australas. Plant Pathol. 1988, 17, 22–23. [Google Scholar] [CrossRef]
- De Brito, N.M.; Duarte, H.S.S.; Bühner, C. de B.; Auer, C.G.; Santos, Á.F. dos. Morphophysiological Characterization of Ceratocystis Fimbriata Isolates from Yerba Mate. Cienc. Rural 2021, 51. [Google Scholar] [CrossRef]
- De Brito, N.M.; Duarte, H.d.S.S.; Auer, C.G.; Wendling, I.; Alfenas, R.F.; Dos Santos, Á.F. Aggressiveness and Screening for Host Responses in Yerba Mate (Ilex Paraguariensis) Clones to a Wilt Disease Caused by Ceratocystis Fimbriata. For. Pathol. 2021, 51, e12682. [Google Scholar] [CrossRef]
- Engelbrecht, C.J.; Harrington, T.C.; Alfenas, A. Ceratocystis Wilt of Cacao—A Disease of Increasing Importance. Phytopathology 2007, 97, 1648–1649. [Google Scholar] [CrossRef] [PubMed]
- Engelbrecht, C.J.B.; Harrington, T.C.; Alfenas, A.C.; Suarez, C. Genetic Variation in Populations of the Cacao Wilt Pathogen, Ceratocystis Cacaofunesta. Plant Pathol. 2007, 56, 923–933. [Google Scholar] [CrossRef]
- Suzuki, N.; Ghabrial, S.A.; Kim, K.-H.; Pearson, M.; Marzano, S.-Y.L.; Yaegashi, H.; Xie, J.; Guo, L.; Kondo, H.; Koloniuk, I.; et al. ICTV Virus Taxonomy Profile: Hypoviridae. J. Gen. Virol. 2018, 99, 615–616. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bian, R.; Liu, Q.; Yang, L.; Pang, T.; Salaipeth, L.; Andika, I.B.; Kondo, H.; Sun, L. Identification of a Novel Hypovirulence-Inducing Hypovirus From Alternaria Alternata. Front. Microbiol. 2019, 10, 1076. [Google Scholar] [CrossRef] [PubMed]
- De Miccolis Angelini, R.M.; Raguseo, C.; Rotolo, C.; Gerin, D.; Faretra, F.; Pollastro, S. The Mycovirome in a Worldwide Collection of the Brown Rot Fungus Monilinia Fructicola. J. Fungi 2022, 8, 481. [Google Scholar] [CrossRef]
- Velasco, L.; Arjona-Girona, I.; Ariza-Fernández, M.T.; Cretazzo, E.; López-Herrera, C. A Novel Hypovirus Species From Xylariaceae Fungi Infecting Avocado. Front. Microbiol. 2018, 9, 778. [Google Scholar] [CrossRef]
- Li, Q.; Huang, W.; Hai, D.; Wang, Y.; Xie, J.; Wang, M. The Complete Genome Sequence of a Novel Hypovirus Infecting Bipolaris Oryzae. Arch. Virol. 2020, 165, 1027–1031. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, J.; Nzabanita, C.; Zhang, M.; Nie, J.; Guo, L. Fungal Virus, FgHV1-Encoded P20 Suppresses RNA Silencing through Single-Strand Small RNA Binding. J. Fungi 2022, 8, 1171. [Google Scholar] [CrossRef]
- Aulia, A.; Hyodo, K.; Hisano, S.; Kondo, H.; Hillman, B.I.; Suzuki, N. Identification of an RNA Silencing Suppressor Encoded by a Symptomless Fungal Hypovirus, Cryphonectria Hypovirus 4. Biology 2021, 10, 100. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Agbandje-McKenna, M.; Canuti, M.; Chiorini, J.A.; Eis-Hubinger, A.-M.; Hughes, J.; Mietzsch, M.; Modha, S.; Ogliastro, M.; Pénzes, J.J.; et al. ICTV Virus Taxonomy Profile: Parvoviridae. J. Gen. Virol. 2019, 100, 367–368. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Q.; Li, S.; Sossah, F.L.; Han, X.; Zhu, G.; Li, Y.; Li, C.; Fu, Y. A Novel Fungal Negative-Stranded RNA Virus Related to Mymonaviruses in Auricularia Heimuer. Arch. Virol. 2022, 167, 2223–2227. [Google Scholar] [CrossRef]
- Hao, F.; Wu, M.; Li, G. Molecular Characterization and Geographic Distribution of a Mymonavirus in the Population of Botrytis Cinerea. Viruses 2018, 10, 432. [Google Scholar] [CrossRef] [PubMed]
- Vainio, E.J.; Chiba, S.; Ghabrial, S.A.; Maiss, E.; Roossinck, M.; Sabanadzovic, S.; Suzuki, N.; Xie, J.; Nibert, M. ICTV Virus Taxonomy Profile: Partitiviridae. J. Gen. Virol. 2018, 99, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, B.; Liu, J.; Wang, Q.; Zhang, Q.; Yu, W.; Hsiang, T. A Novel Mycovirus Infecting Aspergillus Nidulans That Is Closely Related to Viruses in a New Genus of the Family Partitiviridae. Arch. Virol. 2021, 166, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, H.; Kotta-Loizou, I.; Kim, K.-H.; Xie, J. Editorial: Mycoviruses and Related Viruses Infecting Fungi, Lower Eukaryotes, Plants and Insects. Front. Microbiol. 2021, 12, 798598. [Google Scholar] [CrossRef]
- Deng, F.; Boland, G.J. Natural Occurrence of a Partitivirus in the Sapstaining Fungus Ceratocystis Resinifera. Can. J. Plant Pathol. 2007, 29, 182–189. [Google Scholar] [CrossRef]
- Botella, L.; Jung, M.H.; Rost, M.; Jung, T. Natural Populations from the Phytophthora Palustris Complex Show a High Diversity and Abundance of SsRNA and DsRNA Viruses. J. Fungi 2022, 8, 1118. [Google Scholar] [CrossRef]
- Kinsella, C.M.; Deijs, M.; Gittelbauer, H.; van der Hoek, L.; van Dijk, K. Human Clinical Isolates of Pathogenic Fungi Are Host to Diverse Mycoviruses. Microbiol. Spectr. 2022, 10, e01610–e01622. [Google Scholar] [CrossRef]
- Zou, Q.; Gao, Y.; Wang, Q.; Yang, Y.; Wang, F.; Hong, N.; Wang, G.; Wang, L. The Full-Length Genome Sequence of a Novel Mitovirus from Botryosphaeria Dothidea, the Causal Agent of Pear Ring Rot Disease. Arch. Virol. 2021, 166, 2881–2885. [Google Scholar] [CrossRef]
- Wang, H.; Liu, H.; Zhou, Q. The Complete Genome Sequence of a New Mitovirus from the Phytopathogenic Fungus Colletotrichum Higginsianum. Arch. Virol. 2021, 166, 1481–1484. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ni, Y.; Liu, X.; Zhao, H.; Xiao, Y.; Xiao, X.; Li, S.; Liu, H. Divergent RNA Viruses in Macrophomina Phaseolina Exhibit Potential as Virocontrol Agents. Virus Evol. 2021, 7, veaa095. [Google Scholar] [CrossRef] [PubMed]
- de Rezende, R.R.; de Oliveira Souza, F.; Leal, L.L.; Morgan, T.; Bermudez, J.M.M.; Cascardo, R.d.S.; de Barros, D.R.; Alfenas-Zerbini, P. Characterization of a New Mitovirus Infecting the Phytopathogenic Fungus Microdochium Albescens. Arch. Virol. 2021, 166, 3455–3459. [Google Scholar] [CrossRef] [PubMed]
- Shafik, K.; Umer, M.; You, H.; Aboushedida, H.; Wang, Z.; Ni, D.; Xu, W. Characterization of a Novel Mitovirus Infecting Melanconiella Theae Isolated From Tea Plants. Front Microbiol. 2021, 12, 757556. [Google Scholar] [CrossRef]
- Koonin, E.V.; Dolja, V.V.; Morris, T.J. Evolution and Taxonomy of Positive-Strand RNA Viruses: Implications of Comparative Analysis of Amino Acid Sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef]
- Nishikiori, M.; Sugiyama, S.; Xiang, H.; Niiyama, M.; Ishibashi, K.; Inoue, T.; Ishikawa, M.; Matsumura, H.; Katoh, E. Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus. J. Virol. 2012, 86, 7565–7576. [Google Scholar] [CrossRef]
- Luo, X.; Jiang, D.; Xie, J.; Jia, J.; Duan, J.; Cheng, J.; Fu, Y.; Chen, T.; Yu, X.; Li, B.; et al. Genome Characterization and Phylogenetic Analysis of a Novel Endornavirus That Infects Fungal Pathogen Sclerotinia Sclerotiorum. Viruses 2022, 14, 456. [Google Scholar] [CrossRef]
- Cao, C.T.H.; Derbyshire, M.C.; Regmi, R.; Li, H.; Jones, M.G.K.; Wylie, S.J. Small RNA Analyses of a Ceratobasidium Isolate Infected with Three Endornaviruses. Viruses 2022, 14, 2276. [Google Scholar] [CrossRef]
- Marais, A.; Faure, C.; Comont, G.; Candresse, T.; Stempien, E.; Corio-Costet, M.-F. Characterization of the Mycovirome of the Phytopathogenic Fungus, Neofusicoccum Parvum. Viruses 2021, 13, 375. [Google Scholar] [CrossRef]
- Koonin, E.V.; Gorbalenya, A.E.; Chumakov, K.M. Tentative Identification of RNA-Dependent RNA Polymerases of DsRNA Viruses and Their Relationship to Positive Strand RNA Viral Polymerases. FEBS Lett. 1989, 252, 42–46. [Google Scholar] [CrossRef]
- Zanotto, P.M.; Gibbs, M.J.; Gould, E.A.; Holmes, E.C. A Reevaluation of the Higher Taxonomy of Viruses Based on RNA Polymerases. J. Virol. 1996, 70, 6083–6096. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Nuss, D.L. A Host Dicer Is Required for Defective Viral RNA Production and Recombinant Virus Vector RNA Instability for a Positive Sense RNA Virus. Proc. Natl. Acad. Sci. USA 2008, 105, 16749–16754. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Eusebio-Cope, A.; Miyashita, S.; Yokoyama, A.; Aulia, A.; Shahi, S.; Kondo, H.; Suzuki, N. Establishment of Neurospora Crassa as a Model Organism for Fungal Virology. Nat. Commun. 2020, 11, 5627. [Google Scholar] [CrossRef] [PubMed]
- Maiti, M.; Lee, H.-C.; Liu, Y. QIP, a Putative Exonuclease, Interacts with the Neurospora Argonaute Protein and Facilitates Conversion of Duplex SiRNA into Single Strands. Genes Dev. 2007, 21, 590–600. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Liao, X.L.; Gao, B.; Lu, X.; Sun, D.; Gong, W.; Zhong, J.; Zhu, H.; Pan, X.; et al. Mycoviral Gene Integration Converts a Plant Pathogenic Fungus into a Biocontrol Agent. Proc. Natl. Acad. Sci. USA 2022, 119, e2214096119. [Google Scholar] [CrossRef]
- Lee, H.-C.; Chang, S.-S.; Choudhary, S.; Aalto, A.P.; Maiti, M.; Bamford, D.H.; Liu, Y. QiRNA Is a New Type of Small Interfering RNA Induced by DNA Damage. Nature 2009, 459, 274–277. [Google Scholar] [CrossRef]
- de Beer, Z.W.; Duong, T.A.; Barnes, I.; Wingfield, B.D.; Wingfield, M.J. Redefining Ceratocystis and Allied Genera. Stud. Mycol 2014, 79, 187–219. [Google Scholar] [CrossRef]
- Nasution, A.; Glen, M.; Beadle, C.; Mohammed, C. Ceratocystis Wilt and Canker—A Disease That Compromises the Growing of Commercial Acacia-Based Plantations in the Tropics. Aust. For. 2019, 82 (Suppl. S1), 80–93. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).