Role of Gut Microbiota in Breast Cancer and Drug Resistance
Abstract
1. Introduction
2. Microbiome and Breast Cancer, the Connection
3. The Microbiota of Breast and Breast Tumor
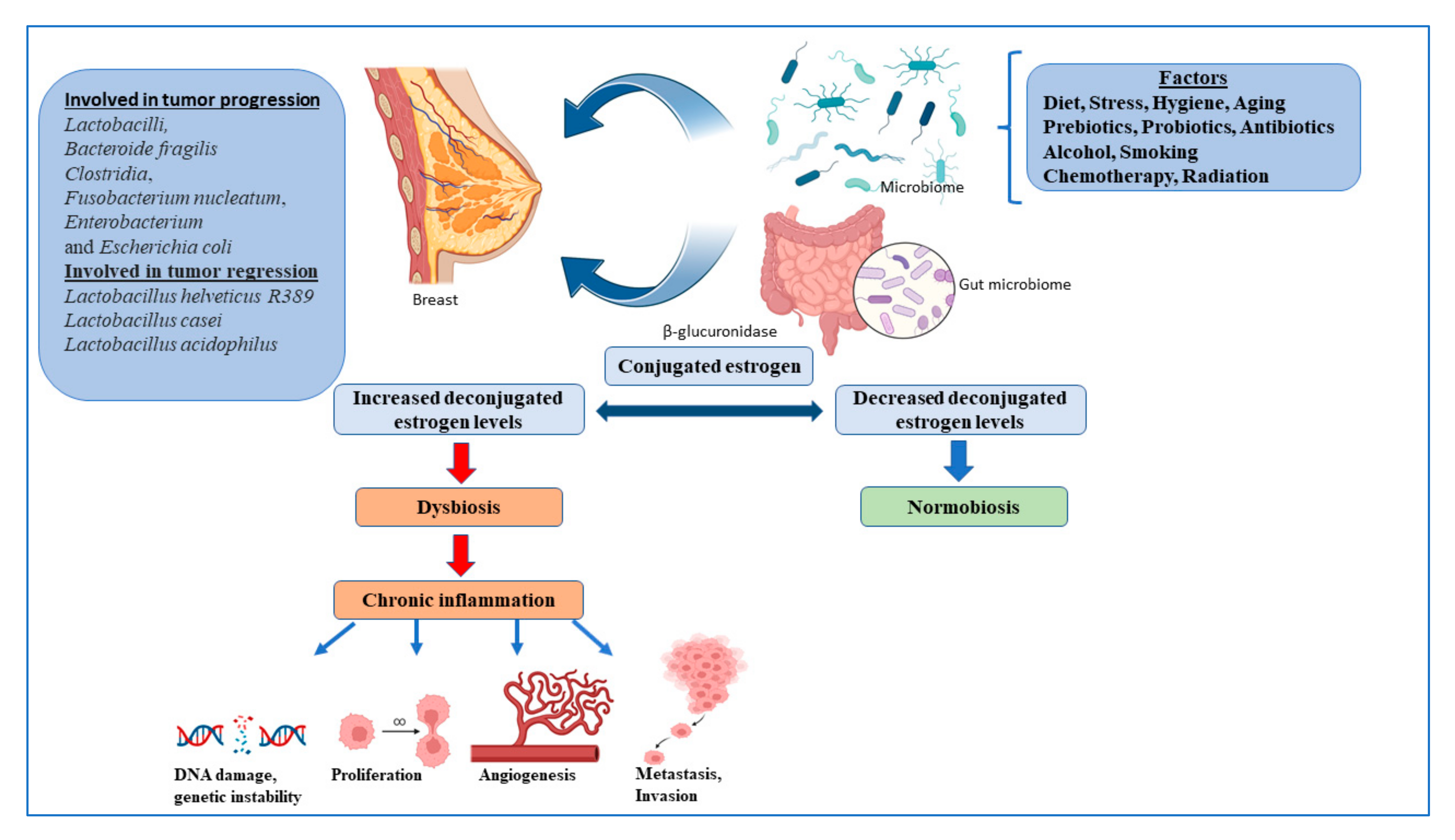
4. Gut Microbiome and Breast Cancer
5. Gut Microbiome and Hormone Therapy
6. Microbiome and Cancer Immunotherapy
7. Microbiota Role in Drug Resistance
8. Clinical Trials-Microbiota and Breast Cancer
9. Microbiota as a Potential Biomarker in Breast Cancer
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BFT | Bacteroides fragilis toxin |
| HBV | Hepatitis B virus |
| BCV | Hepatitis C virus |
| ER | Estrogen receptor |
| PR | progesterone receptor |
| TNBC | Triple negative breast cancer |
| HMP | Human microbiome project |
| iHMP | Integrated Human microbiome project |
| IDC | Invasive ductal carcinoma |
| ILC | Invasive ductal carcinoma |
| PAMPs | Pathogen-associated molecular patterns |
| ICIS | Immune checkpoint inhibitors |
| T-regs | Regulatory T cells |
| FMT | Fecal microbial transplantation |
| pCR | Pathological complete response |
| OTUs | Operational Taxonomic Units |
| PCOS | Polycystic ovary syndrome |
| CVD | Cardiovascular disease |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Lacey, J.V., Jr.; Kreimer, A.R.; Buys, S.S.; Marcus, P.M.; Chang, S.C.; Leitzmann, M.F.; Hoover, R.N.; Prorok, P.C.; Berg, C.D.; Hartge, P.; et al. Breast cancer epidemiology according to recognized breast cancer risk factors in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial Cohort. BMC Cancer 2009, 9, 84. [Google Scholar] [CrossRef] [PubMed]
- Morales-Sanchez, A.; Fuentes-Panana, E.M. Human viruses and cancer. Viruses 2014, 6, 4047–4079. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Blaser, M.J.; Caporaso, J.G.; Jansson, J.K.; Lynch, S.V.; Knight, R. Current understanding of the human microbiome. Nat. Med. 2018, 24, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.V.; Pedersen, O. The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 2016, 375, 2369–2379. [Google Scholar] [CrossRef]
- Allen, J.; Sears, C.L. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: Contributions to colorectal cancer development. Genome Med. 2019, 11, 11. [Google Scholar] [CrossRef]
- Kadosh, E.; Snir-Alkalay, I.; Venkatachalam, A.; May, S.; Lasry, A.; Elyada, E.; Zinger, A.; Shaham, M.; Vaalani, G.; Mernberger, M.; et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020, 586, 133–138. [Google Scholar] [CrossRef]
- Parida, S.; Wu, S.; Siddharth, S.; Wang, G.; Muniraj, N.; Nagalingam, A.; Hum, C.; Mistriotis, P.; Hao, H.; Talbot, C.C., Jr.; et al. A Procarcinogenic Colon Microbe Promotes Breast Tumorigenesis and Metastatic Progression and Concomitantly Activates Notch and beta-Catenin Axes. Cancer Discov. 2021, 11, 1138–1157. [Google Scholar] [CrossRef]
- Hwang, S.; Yi, H.C.; Hwang, S.; Jo, M.; Rhee, K.J. Dietary Salt Administration Decreases Enterotoxigenic Bacteroides fragilis (ETBF)-Promoted Tumorigenesis via Inhibition of Colonic Inflammation. Int. J. Mol. Sci. 2020, 21, 8034. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Tian, Z.; Kong, X.; Yang, L.; Shan, X.; Dong, B.; Ding, X.; Jing, X.; Jiang, C.; Jiang, N.; et al. FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 2020, 39, 202. [Google Scholar] [CrossRef]
- Mellemgaard, A.; Gaarslev, K. Risk of hepatobiliary cancer in carriers of Salmonella typhi. J. Natl. Cancer Inst. 1988, 80, 288. [Google Scholar] [CrossRef] [PubMed]
- Oliero, M.; Hajjar, R.; Cuisiniere, T.; Fragoso, G.; Calve, A.; Dagbert, F.; Loungnarath, R.; Sebajang, H.; Schwenter, F.; Wassef, R.; et al. Prevalence of pks + bacteria and enterotoxigenic Bacteroides fragilis in patients with colorectal cancer. Gut Pathog. 2022, 14, 51. [Google Scholar] [CrossRef]
- Grivennikov, S.I. IL-11: A prominent pro-tumorigenic member of the IL-6 family. Cancer Cell 2013, 24, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Fulbright, L.E.; Ellermann, M.; Arthur, J.C. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017, 13, e1006480. [Google Scholar] [CrossRef]
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef]
- The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.; Sharma, D. The power of small changes: Comprehensive analyses of microbial dysbiosis in breast cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1871, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Pierre, J.F.; Makowski, L.; Tolley, E.; Lyn-Cook, B.; Lu, L.; Vidal, G.; Starlard-Davenport, A. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 2019, 9, 11940. [Google Scholar] [CrossRef]
- Tzeng, A.; Sangwan, N.; Jia, M.; Liu, C.C.; Keslar, K.S.; Downs-Kelly, E.; Fairchild, R.L.; Al-Hilli, Z.; Grobmyer, S.R.; Eng, C. Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 2021, 13, 60. [Google Scholar] [CrossRef]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N.; et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef]
- He, Z.; Tian, W.; Wei, Q.; Xu, J. Involvement of Fusobacterium nucleatum in malignancies except for colorectal cancer: A literature review. Front. Immunol. 2022, 13, 968649. [Google Scholar] [CrossRef]
- Yazdi, M.H.; Soltan Dallal, M.M.; Hassan, Z.M.; Holakuyee, M.; Agha Amiri, S.; Abolhassani, M.; Mahdavi, M. Oral administration of Lactobacillus acidophilus induces IL-12 production in spleen cell culture of BALB/c mice bearing transplanted breast tumour. Br. J. Nutr. 2010, 104, 227–232. [Google Scholar] [CrossRef]
- De Moreno de LeBlanc, A.; Matar, C.; Theriault, C.; Perdigon, G. Effects of milk fermented by Lactobacillus helveticus R389 on immune cells associated to mammary glands in normal and a breast cancer model. Immunobiology 2005, 210, 349–358. [Google Scholar] [CrossRef]
- Soltan Dallal, M.M.; Yazdi, M.H.; Holakuyee, M.; Hassan, Z.M.; Abolhassani, M.; Mahdavi, M. Lactobacillus casei ssp.casei induced Th1 cytokine profile and natural killer cells activity in invasive ductal carcinoma bearing mice. Iran. J. Allergy Asthma Immunol. 2012, 11, 183–189. [Google Scholar]
- Toi, M.; Hirota, S.; Tomotaki, A.; Sato, N.; Hozumi, Y.; Anan, K.; Nagashima, T.; Tokuda, Y.; Masuda, N.; Ohsumi, S.; et al. Probiotic Beverage with Soy Isoflavone Consumption for Breast Cancer Prevention: A Case-control Study. Curr. Nutr. Food Sci. 2013, 9, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e1326. [Google Scholar] [CrossRef]
- Bodai, B.I.; Nakata, T.E. Breast Cancer: Lifestyle, the Human Gut Microbiota/Microbiome, and Survivorship. Perm. J. 2020, 24. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells 2020, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, T.; Miko, E.; Vida, A.; Sebo, E.; Toth, J.; Csonka, T.; Boratko, A.; Ujlaki, G.; Lente, G.; Kovacs, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef]
- Sari, Z.; Miko, E.; Kovacs, T.; Boratko, A.; Ujlaki, G.; Janko, L.; Kiss, B.; Uray, K.; Bai, P. Indoxylsulfate, a Metabolite of the Microbiome, Has Cytostatic Effects in Breast Cancer via Activation of AHR and PXR Receptors and Induction of Oxidative Stress. Cancers 2020, 12, 2915. [Google Scholar] [CrossRef]
- Kovacs, P.; Csonka, T.; Kovacs, T.; Sari, Z.; Ujlaki, G.; Sipos, A.; Karanyi, Z.; Szeocs, D.; Hegedus, C.; Uray, K.; et al. Lithocholic Acid, a Metabolite of the Microbiome, Increases Oxidative Stress in Breast Cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef]
- Parida, S.; Sharma, D. The Microbiome-Estrogen Connection and Breast Cancer Risk. Cells 2019, 8, 1642. [Google Scholar] [CrossRef]
- Vitorino, M.; Baptista de Almeida, S.; Alpuim Costa, D.; Faria, A.; Calhau, C.; Azambuja Braga, S. Human Microbiota and Immunotherapy in Breast Cancer—A Review of Recent Developments. Front. Oncol. 2021, 11, 815772. [Google Scholar] [CrossRef]
- Rea, D.; Coppola, G.; Palma, G.; Barbieri, A.; Luciano, A.; Del Prete, P.; Rossetti, S.; Berretta, M.; Facchini, G.; Perdona, S.; et al. Microbiota effects on cancer: From risks to therapies. Oncotarget 2018, 9, 17915–17927. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Ni, Q.; Sun, W.; Li, L.; Feng, X. The links between gut microbiota and obesity and obesity related diseases. Biomed. Pharmacother. 2022, 147, 112678. [Google Scholar] [CrossRef] [PubMed]
- Eslami, S.Z.; Majidzadeh, A.K.; Halvaei, S.; Babapirali, F.; Esmaeili, R. Microbiome and Breast Cancer: New Role for an Ancient Population. Front. Oncol. 2020, 10, 120. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.G. The relationship of estrogen and progesterone to breast disease. J. Reprod. Med. 1984, 29, 530–538. [Google Scholar]
- Jerry, D.J. Roles for estrogen and progesterone in breast cancer prevention. Breast Cancer Res. 2007, 9, 102. [Google Scholar] [CrossRef]
- Truin, W.; Roumen, R.M.H.; Siesling, S.; van de Vijver, K.K.; Tjan-Heijnen, V.C.G.; Voogd, A.C. Estrogen and progesterone receptor expression levels do not differ between lobular and ductal carcinoma in patients with hormone receptor-positive tumors. Breast Cancer Res. Treat. 2017, 164, 133–138. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Yamamoto, S.; Sobue, T.; Kobayashi, M.; Sasaki, S.; Tsugane, S.; Japan Public Health Center-Based Prospective Study on Cancer Cardiovascular Diseases (JPHC Study) Group. Soy, isoflavones, and breast cancer risk in Japan. J. Natl. Cancer Inst. 2003, 95, 906–913. [Google Scholar] [CrossRef]
- Wada, K.; Nakamura, K.; Tamai, Y.; Tsuji, M.; Kawachi, T.; Hori, A.; Takeyama, N.; Tanabashi, S.; Matsushita, S.; Tokimitsu, N.; et al. Soy isoflavone intake and breast cancer risk in Japan: From the Takayama study. Int. J. Cancer 2013, 133, 952–960. [Google Scholar] [CrossRef]
- Katherine, L.C. Probiotic Bacteria May Enhance Tamoxifen Effectiveness in Treatment of ER+ Breast Cancer; Press Release; Endocrine Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Li, W.; Deng, Y.; Chu, Q.; Zhang, P. Gut microbiome and cancer immunotherapy. Cancer Lett. 2019, 447, 41–47. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Killock, D. Immunotherapy: Gut bacteria modulate responses to PD-1 blockade. Nat. Rev. Clin. Oncol. 2018, 15, 6–7. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Kim, G.; Jeon, B.N.; Fang, S.; Park, H. Bifidobacterium Strain-Specific Enhances the Efficacy of Cancer Therapeutics in Tumor-Bearing Mice. Cancers 2021, 13, 957. [Google Scholar] [CrossRef] [PubMed]
- Toth, G.; Szollosi, J.; Abken, H.; Vereb, G.; Szoor, A. A Small Number of HER2 Redirected CAR T Cells Significantly Improves Immune Response of Adoptively Transferred Mouse Lymphocytes against Human Breast Cancer Xenografts. Int. J. Mol. Sci. 2020, 21, 1039. [Google Scholar] [CrossRef]
- Szoor, A.; Toth, G.; Zsebik, B.; Szabo, V.; Eshhar, Z.; Abken, H.; Vereb, G. Trastuzumab derived HER2-specific CARs for the treatment of trastuzumab-resistant breast cancer: CAR T cells penetrate and eradicate tumors that are not accessible to antibodies. Cancer Lett. 2020, 484, 1–8. [Google Scholar] [CrossRef]
- Sun, M.; Shi, H.; Liu, C.; Liu, J.; Liu, X.; Sun, Y. Construction and evaluation of a novel humanized HER2-specific chimeric receptor. Breast Cancer Res. 2014, 16, R61. [Google Scholar] [CrossRef]
- Priceman, S.J.; Tilakawardane, D.; Jeang, B.; Aguilar, B.; Murad, J.P.; Park, A.K.; Chang, W.C.; Ostberg, J.R.; Neman, J.; Jandial, R.; et al. Regional Delivery of Chimeric Antigen Receptor-Engineered T Cells Effectively Targets HER2(+) Breast Cancer Metastasis to the Brain. Clin. Cancer Res. 2018, 24, 95–105. [Google Scholar] [CrossRef]
- Xia, L.; Zheng, Z.; Liu, J.Y.; Chen, Y.J.; Ding, J.; Hu, G.S.; Hu, Y.H.; Liu, S.; Luo, W.X.; Xia, N.S.; et al. Targeting Triple-Negative Breast Cancer with Combination Therapy of EGFR CAR T Cells and CDK7 Inhibition. Cancer Immunol. Res. 2021, 9, 707–722. [Google Scholar] [CrossRef]
- Xia, L.; Zheng, Z.Z.; Liu, J.Y.; Chen, Y.J.; Ding, J.C.; Xia, N.S.; Luo, W.X.; Liu, W. EGFR-targeted CAR-T cells are potent and specific in suppressing triple-negative breast cancer both in vitro and in vivo. Clin. Transl. Immunol. 2020, 9, e01135. [Google Scholar] [CrossRef]
- Hu, Z. Tissue factor as a new target for CAR-NK cell immunotherapy of triple-negative breast cancer. Sci. Rep. 2020, 10, 2815. [Google Scholar] [CrossRef]
- Makena, M.R.; Ranjan, A.; Thirumala, V.; Reddy, A.P. Cancer stem cells: Road to therapeutic resistance and strategies to overcome resistance. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165339. [Google Scholar] [CrossRef] [PubMed]
- Shiao, S.L.; Kershaw, K.M.; Limon, J.J.; You, S.; Yoon, J.; Ko, E.Y.; Guarnerio, J.; Potdar, A.A.; McGovern, D.P.B.; Bose, S.; et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell 2021, 39, 1202–1213.e1206. [Google Scholar] [CrossRef] [PubMed]
- Daillere, R.; Vetizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.M.; Flament, C.; Lepage, P.; Roberti, M.P.; et al. Enterococcus hirae and Barnesiella intestinihominis Facilitate Cyclophosphamide-Induced Therapeutic Immunomodulatory Effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Dieleman, S.; Aarnoutse, R.; Ziemons, J.; Kooreman, L.; Boleij, A.; Smidt, M. Exploring the Potential of Breast Microbiota as Biomarker for Breast Cancer and Therapeutic Response. Am. J. Pathol. 2021, 191, 968–982. [Google Scholar] [CrossRef]
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacol. Res. 2019, 148, 104398. [Google Scholar] [CrossRef]
- Kang, M.H.; Wang, J.; Makena, M.R.; Lee, J.S.; Paz, N.; Hall, C.P.; Song, M.M.; Calderon, R.I.; Cruz, R.E.; Hindle, A.; et al. Activity of MM-398, nanoliposomal irinotecan (nal-IRI), in Ewing’s family tumor xenografts is associated with high exposure of tumor to drug and high SLFN11 expression. Clin. Cancer Res. 2015, 21, 1139–1150. [Google Scholar] [CrossRef]
- Guthrie, L.; Gupta, S.; Daily, J.; Kelly, L. Human microbiome signatures of differential colorectal cancer drug metabolism. NPJ Biofilms Microbiomes 2017, 3, 27. [Google Scholar] [CrossRef]
- Di Modica, M.; Gargari, G.; Regondi, V.; Bonizzi, A.; Arioli, S.; Belmonte, B.; De Cecco, L.; Fasano, E.; Bianchi, F.; Bertolotti, A.; et al. Gut Microbiota Condition the Therapeutic Efficacy of Trastuzumab in HER2-Positive Breast Cancer. Cancer Res. 2021, 81, 2195–2206. [Google Scholar] [CrossRef] [PubMed]
- Kelley, S.T.; Skarra, D.V.; Rivera, A.J.; Thackray, V.G. The Gut Microbiome Is Altered in a Letrozole-Induced Mouse Model of Polycystic Ovary Syndrome. PLoS ONE 2016, 11, e0146509. [Google Scholar] [CrossRef] [PubMed]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Goedert, J.J.; Hua, X.; Bielecka, A.; Okayasu, I.; Milne, G.L.; Jones, G.S.; Fujiwara, M.; Sinha, R.; Wan, Y.; Xu, X.; et al. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br. J. Cancer 2018, 118, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef]
| Title | Clinical Trial No. | Study Design | Status |
|---|---|---|---|
| Gut Microbiome Components Predict Response to Neoadjuvant Therapy in HER2-positive Breast Cancer Patients: A Prospective Study | NCT05444647 | Observational model: Cohort | Recruiting |
| Exercise, Gut Microbiome, and Breast Cancer: Increasing Reach to Underserved Populations (EMBRACE) | NCT05000502 | Randomized | Recruiting |
| Assessing the Impact of the Microbiome on Breast Cancer Radiotherapy Toxicity | NCT04245150 | Observational model: Cohort | Recruiting |
| Gut and Intratumoral Microbiome Effect on the Neoadjuvant Chemotherapy-induced Immunosurveillance in Triple Negative Breast Cancer | NCT03586297 | Observational model: Cohort | Recruiting |
| The Association Between Radiation Dermatitis and Skin Microbiome in Breast Cancer Patients | NCT05032768 | Observational model: Cohort | Recruiting |
| Engineering Gut Microbiome to Target Breast Cancer | NCT03358511 | Intervention model: Single group assignment | Completed |
| Evaluating the Association Between Changes in the Gut Microbiome and Chemotherapy-Induced Nausea in Women Receiving Chemotherapy for Stage I-III Breast Cancer | NCT05417867 | Observational model: Case-only | Recruiting |
| Evaluating Mepitel in Post-mastectomy Patients and the Role of the Skin Microbiome in Radiation Dermatitis | NCT03519438 | Observational model: Cohort | Completed |
| Determinants of Acquired Endocrine Resistance in Metastatic Breast Cancer: A Pilot Study (ENDO-RESIST) | NCT04579484 | Observational model: Cohort | Recruiting |
| Oral Aromatase Inhibitors Modify the Gut Microbiome | NCT05030038 | Observational model: Cohort | Recruiting |
| The Breast Cancer Personalized Nutrition Study (BREACPNT) | NCT04079270 | Interventional: Randomized | Recruiting |
| Microbiome and Association With Implant Infections | NCT05020574 | Interventional: Randomized (phase 2) | Recruiting |
| Gut Microbe Composition, Exercise, and Breast Breast Cancer Survivors (ROME) | NCT04088708 | Interventional: Randomized | Recruiting |
| Effect of Radiotherapy Variables on Circulating Effectors of Immune Response and Local Microbiome | NCT03383107 | Observational | Completed |
| Study to Investigate Efficacy of a Novel Probiotic on the Bacteriome and Mycobiome of Breast Cancer | NCT04362826 | Interventional: Randomized | Not yet recruited |
| ARGONAUT: Stool and Blood Sample Bank for Cancer Patients | NCT04638751 | Observational model: Cohort | Recruiting |
| The Gut Microbiome and Immune Checkpoint Inhibitor Therapy in Solid Tumors (PARADIGM) | NCT05037825 | Observational model: Cohort | Recruiting |
| Anti-anxiety Biotics for Breast Cancer Survivors (ABBCS) | NCT04784182 | Interventional: Randomized | Completed |
| Adaptive Nutrition and Exercise Weight Loss (A-NEW) Study (A-NEW) | NCT04499950 | Interventional: non-randomized | Recruiting |
| Effects of Probiotics on the Gut Microbiome and Immune System in Operable Stage I-III Breast or Lung Cancer | NCT04857697 | Interventional | Recruiting |
| Probiotics and Breast Health | NCT03290651 | Interventional | Completed |
| Intestinal Microbiota Impact for Prognosis and Treatment Outcomes in Early Luminal Breast Cancer and Pancreatic Cancer Patients | NCT05580887 | Observational model: Cohort | Recruiting |
| Intestine Bacteria and Breast Cancer Risk | NCT01461070 | Observational model: Case-only | Completed |
| Neoadjuvant Treatment of Locally-advanced Breast Cancer Patients With Ribociclib and Letrozole (NEOLETRIB) | NCT05163106 | Interventional | Recruiting |
| Persistent Post-Surgical Pain in Women With BrCA | NCT02266082 | Observational model: Cohort | Completed |
| Study of Moderate Dose Omega 3 Fatty Acid Supplement in Premenopausal Women at High Risk for Breast Cancer | NCT03383835 | Interventional | Un-Known |
| GRACE-trial: a Randomized Active-controlled Trial for vulvovaginal atrophy in breast Cancer Patients on Endocrine Therapy. (GRACE) | NCT05562518 | Interventional: Randomized | Recruiting |
| Comprehensive Outcomes for After Cancer Health (COACH) | NCT05349227 | Interventional: Randomized | Recruiting |
| Rifaximin for the Treatment of Gastrointestinal Toxicities Related to Pertuzumab-Based Therapy in Patients With Stage I-III HER2 Positive Breast Cancer | NCT04249622 | Interventional: Non-Randomized | Recruiting |
| Impact of Vitamin D Supplementation on the Rate of Pathologic Complete Response in Vitamin D Deficient Patients | NCT04677816 | Interventional: Non-Randomized | Recruiting |
| Weight Loss Plus Omega-3 Fatty Acids or Placebo in High Risk Women | NCT02101970 | Interventional: Randomized | Recruiting |
| Comprehensive Lifestyle Change To Prevent Breast Cancer | NCT03448003 | Interventional: Randomized | Recruiting |
| Avera/Sema4 Oncology and Analytics Protocol (ASAP) | NCT05142033 | Interventional: Randomized | Recruiting |
| Gender Difference in Side effects of Immunotherapy: A Possible Clue to Optimize Cancer Treatment (G-DEFINER) | NCT04435964 | Observational | Recruiting |
| Neoadjuvant Pembrolizumab(Pbr)/Nab-Paclitaxel Followed by Pbr/Epirubicin/Cyclophosphamide in TNBC (NIB) | NCT03289819 | Interventional | Completed |
| Abemaciclib in Treating Patients With Surgically Resectable, Chemotherapy Resistant, Triple Negative Breast Cancer | NCT03979508 | Interventional: Non-Randomized | Recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viswanathan, S.; Parida, S.; Lingipilli, B.T.; Krishnan, R.; Podipireddy, D.R.; Muniraj, N. Role of Gut Microbiota in Breast Cancer and Drug Resistance. Pathogens 2023, 12, 468. https://doi.org/10.3390/pathogens12030468
Viswanathan S, Parida S, Lingipilli BT, Krishnan R, Podipireddy DR, Muniraj N. Role of Gut Microbiota in Breast Cancer and Drug Resistance. Pathogens. 2023; 12(3):468. https://doi.org/10.3390/pathogens12030468
Chicago/Turabian StyleViswanathan, Sathiyapriya, Sheetal Parida, Bhuvana Teja Lingipilli, Ramalingam Krishnan, Devendra Rao Podipireddy, and Nethaji Muniraj. 2023. "Role of Gut Microbiota in Breast Cancer and Drug Resistance" Pathogens 12, no. 3: 468. https://doi.org/10.3390/pathogens12030468
APA StyleViswanathan, S., Parida, S., Lingipilli, B. T., Krishnan, R., Podipireddy, D. R., & Muniraj, N. (2023). Role of Gut Microbiota in Breast Cancer and Drug Resistance. Pathogens, 12(3), 468. https://doi.org/10.3390/pathogens12030468









