Immunoreactivity of a Putative ECF σ Factor, LIC_10559, from Leptospira interrogans with Sera from Leptospira-Infected Animals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Serum Samples
2.2. Plasmid Construction for Protein Overproduction
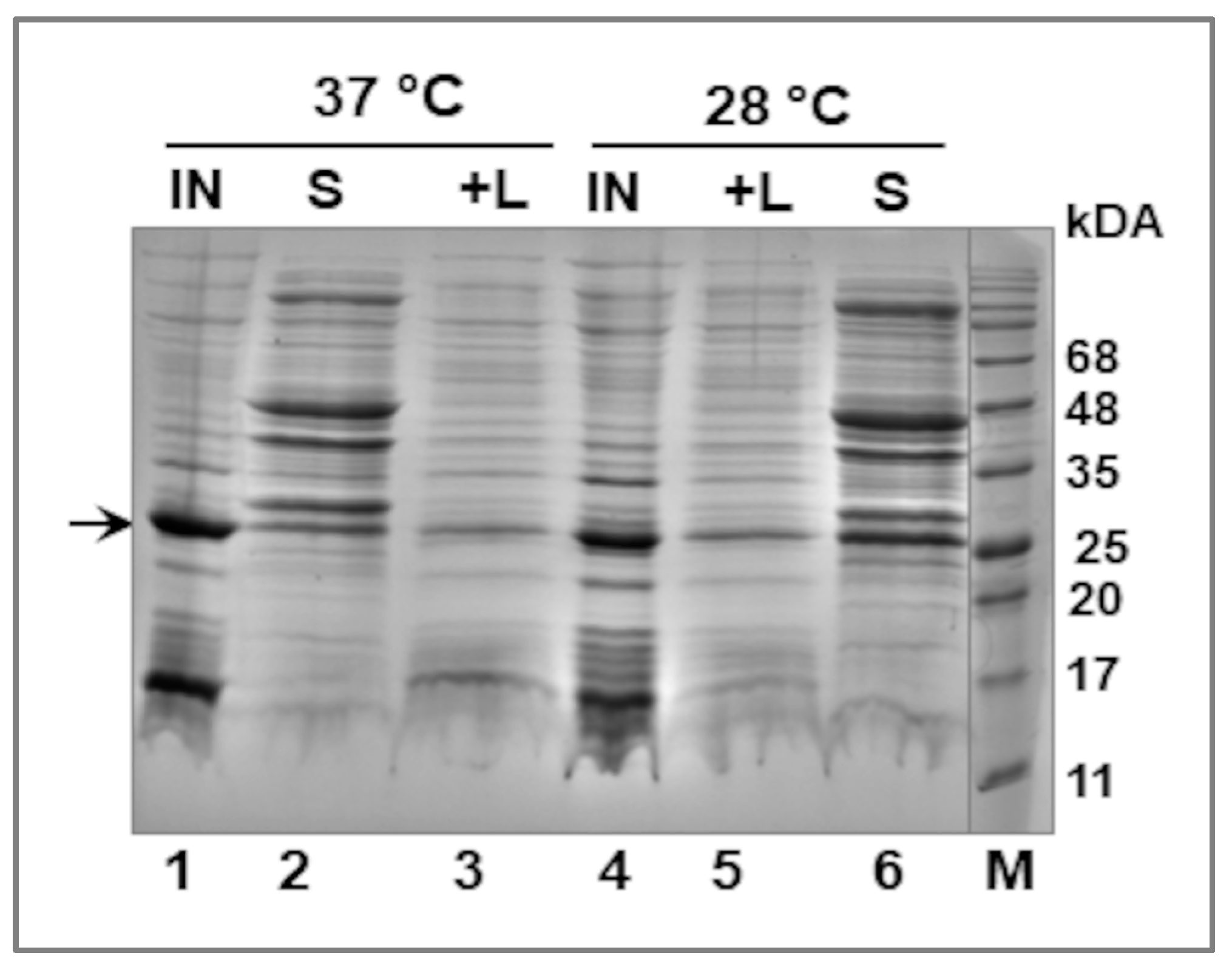
2.3. Fractionation of E. coli Cells—Extraction of the Recombinant LIC_10559 Protein
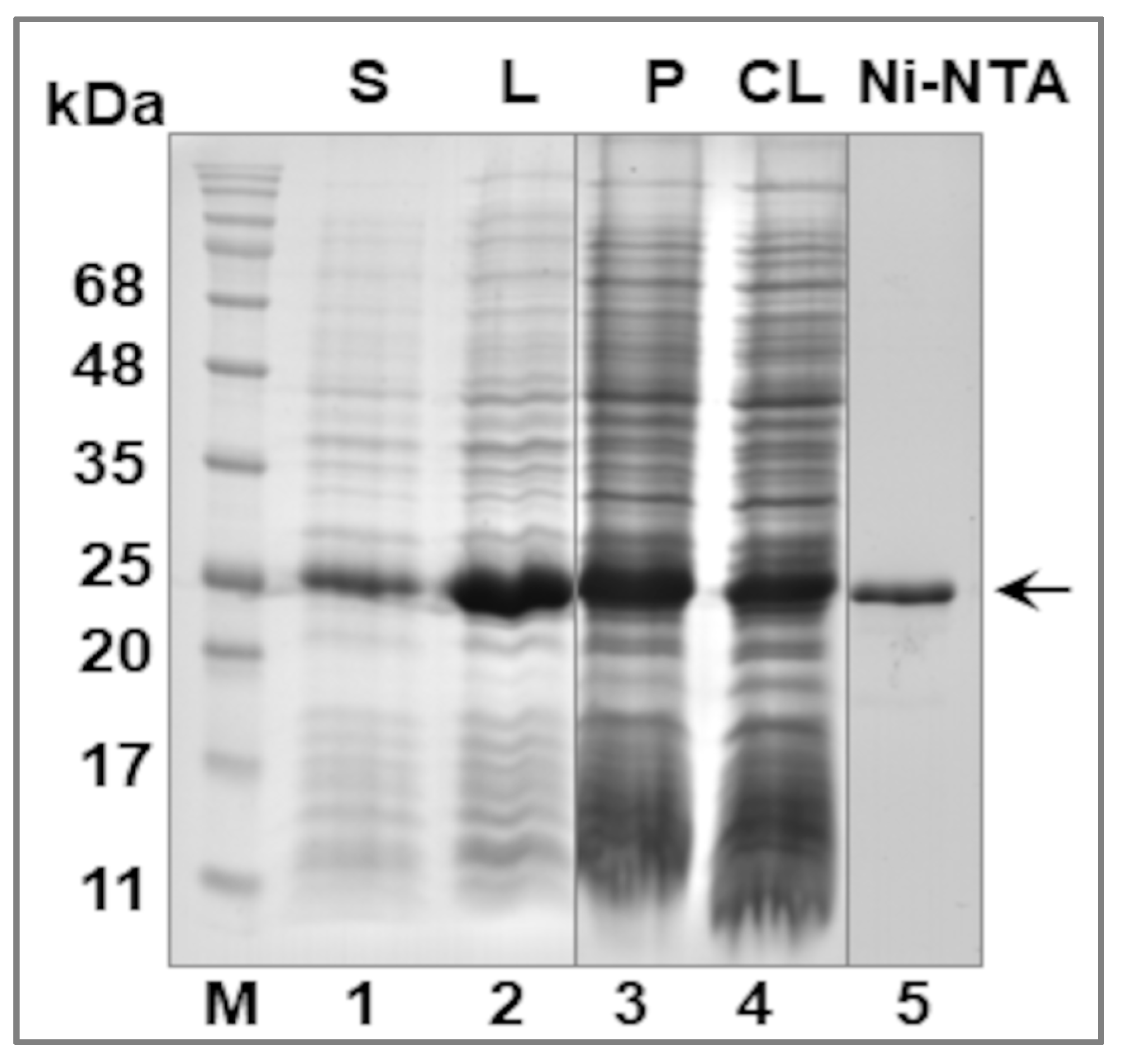
2.4. Purification of the Recombinant LIC_10559 Protein
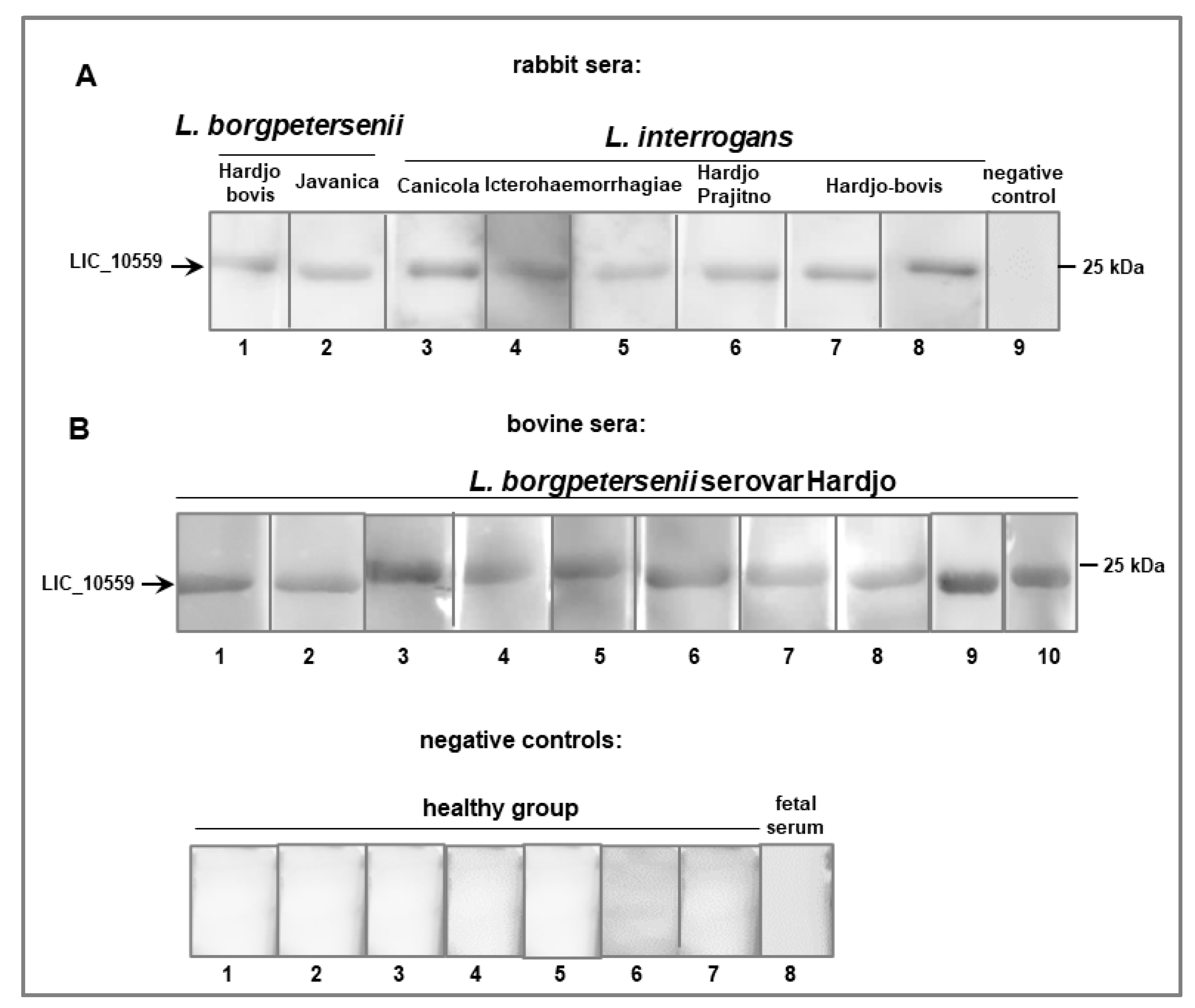
2.5. SDS-PAGE and Western Blotting Analysis
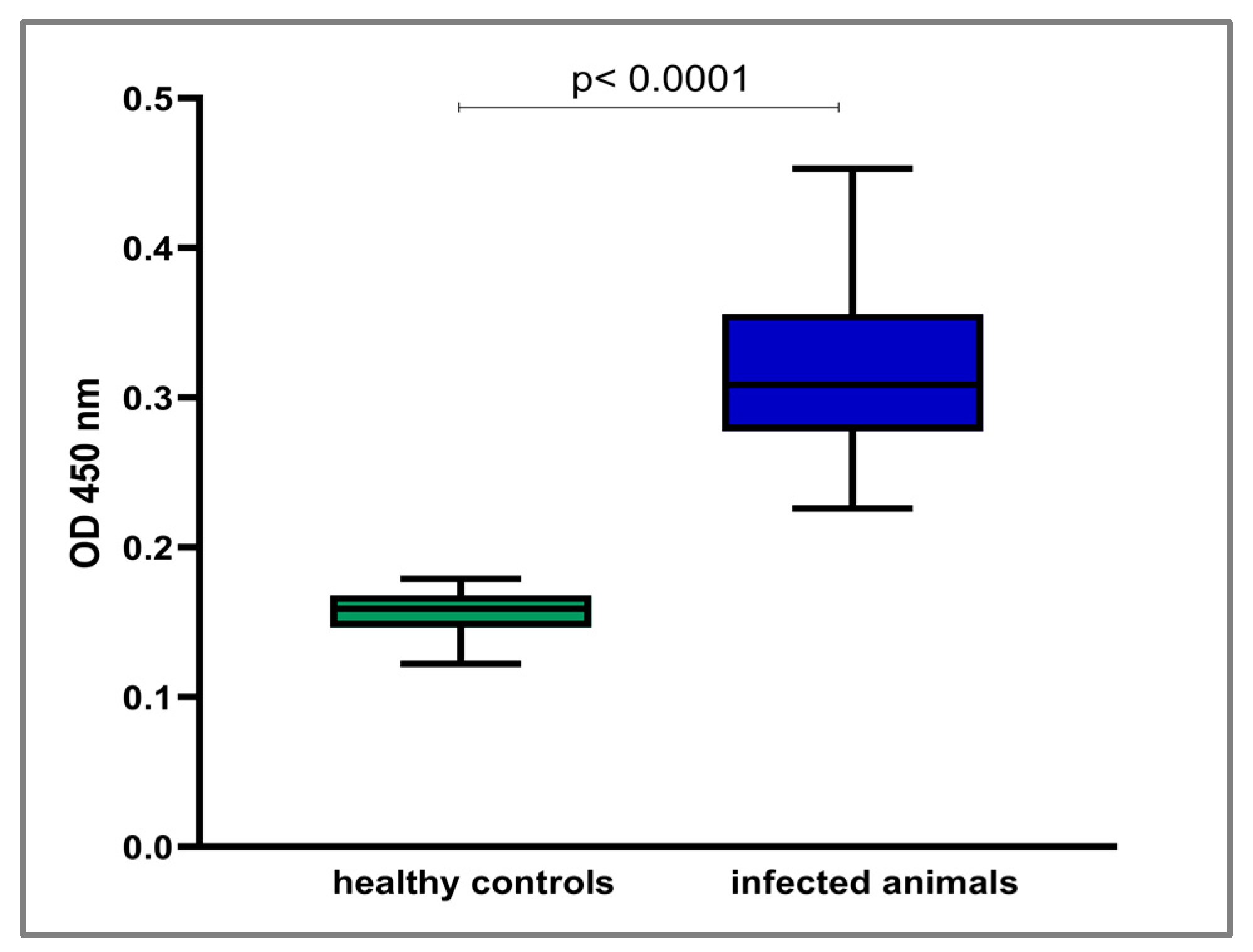
2.6. ELISA Assay
2.7. Data Analysis
3. Results and Discussion
3.1. LIC_10559 Sequence Analysis Using Bioinformatics Tools and Protein Databases
3.2. Expression of LIC_10559 in E. coli pET System and Purification of Its Protein Product
3.3. Immunogenic Potential of LIC_10559
4. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pal, M.; Bulcha, M.R.; Bune, W.M. Leptospirosis and one health perspective. Am. J. Public. Health. Res. 2021, 9, 180–183. [Google Scholar] [CrossRef]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.L. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. Curr. Top. Microbiol. Immunol. 2015, 387, 65–97. [Google Scholar]
- Yanagihara, Y.; Villsnueva, S.Y.A.M.; Nomura, N.; Ohno, M.; Sekiya, T.; Handabile, C.; Shingai, M.; Higashi, H.; Yoshida, S.I.; Masuzawa, T.; et al. Leptospira is an environmental bacterium that grows in waterlogged soil. Microbiol. Spectr. 2022, 10, e0215721. [Google Scholar] [CrossRef] [PubMed]
- Arent, Z.J.; Gilmore, C.; San-Miguel Ayanz, J.M.; Neyra, L.Q.; García-Peña, F.J. Molecular epidemiology of Leptospira serogroup Pomona infections among wild and domestic animals in Spain. EcoHealth 2017, 14, 48–57. [Google Scholar] [CrossRef]
- Pejsak, Z.; Truszczyński, M.; Arent, Z. Leptospirosis in swine in the light of accepted characterization. Med. Wet. 2020, 76, 555–558. [Google Scholar] [CrossRef]
- Arent, Z.; Kędzierska-Mieszkowska, S. Seroprevalence study of leptospirosis in horses in northern Poland. Vet. Rec. 2013, 172, 269. [Google Scholar] [CrossRef]
- Xue, F.; Dong, H.; Wu, J.; Wu, Z.; Hu, W.; Sun, A.; Troxell, B.; Yang, X.F.; Yan, J. Transcriptional responses of Leptospira interrogans to host innate immunity: Significant changes in metabolism, oxygen tolerance, and outer membrane. PLos. Negl. Trop. Dis. 2010, 4, e857. [Google Scholar] [CrossRef] [Green Version]
- Lo, M.; Bulach, D.M.; Powell, D.R.; Haake, D.A.; Matsunaga, J.; Paustian, M.L.; Zuerner, R.L.; Adler, B. Effects of temperature on gene expression patterns in Leptospira interrogans serovar Lai as assessed by whole-genome microarrays. Infect. Immun. 2006, 74, 5848–5859. [Google Scholar] [CrossRef] [Green Version]
- Lo, M.; Murray, G.; Khoo, C.A.; Haake, D.A.; Zuerner, R.L.; Adler, B. Transcriptional response of Leptospira interrogans to iron limitation and characterization of a PerR homolog. Infect. Immun. 2010, 78, 4850–4859. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.H.; Sheng, Y.Y.; Zhang, Z.; Shi, Y.Z.; He, P.; Hu, B.-Y.; Yang, Y.; Liu, S.-G.; Zhao, G.-P.; Guo, X.-K. Genome-wide transcriptional analysis of temperature shift in L. interrogans serovar Lai strain 56601. BMC Microbiol. 2006, 6, 51. [Google Scholar] [CrossRef] [Green Version]
- Matsunaga, J.; Lo, M.; Bulach, D.M.; Zuerner, R.L.; Adler, B.; Haake, D.A. Response of Leptospira interrogans to physiologic osmolarity: Relevance in signaling the environment-to-host transition. Infect. Immun. 2007, 75, 2864–2874. [Google Scholar] [CrossRef] [Green Version]
- Patarakul, K.; Lo, M.; Adler, B. Global transcriptomic response of Leptospira interrogans serovar Copenhageni upon exposure to serum. BMC Microbiol. 2010, 10, 31. [Google Scholar] [CrossRef] [Green Version]
- Caimo, M.J.; Sivasankaran, S.K.; Allard, A.; Hurley, D.; Hokamp, K.; Grassmann, A.A.; Hinton, J.C.; Nally, J.E. A model system for studying the transcriptomic and physiological changes associated with mammalian host-adaptation by Leptospira interrogans serovar Copenhageni. PLoS Pathog. 2014, 10, e100404. [Google Scholar]
- Nally, J.E.; Grassmann, A.A.; Planchon, S.; Sergeant, K.; Renaut, J.; Seshu, J.; McBride, A.J.; Caimano, M. Pathogenic leptospires modulate protein expression and post-translational modifications in response to mammalian host signals. Front. Cell Infect. Microbiol. 2017, 7, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.K.; Dhasmana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial virulence factors: Secreted for survival. Indian. J. Microbiol. 2017, 57, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foust, D.E.; Matthias, M.A.; Adhikarla, H.; Adler, B.; Amorim-Santos, L.; Berg, D.; Bulach, D.; Buschiazzo, A.; Chang, Y.-F.; Galloway, R.L.; et al. What makes a bacterial species pathogenic? Comparative gnomic analyses of the genus Leptospira. PLoS Negl. Trop. Dis. 2016, 10, e0004403. [Google Scholar] [CrossRef] [Green Version]
- Zhukova, A.; Fernandes, L.G.; Hugon, P.; Pappas, C.J.; Sismeiro, O.; Coppée, J.Y.; Becavin, C.; Malabat, C.; Eshghi, A.; Zhang, J.J.; et al. Picardeau M. Genome-Wide Transcriptional Start Site Mapping and sRNA Identification in the Pathogen Leptospira interrogans. Front. Cell. Infect. Microbiol. 2017, 7, 10. [Google Scholar] [CrossRef] [Green Version]
- Helmann, J.D. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002, 46, 47–110. [Google Scholar]
- Manganelli, R.; Fattorini, L.; Tan, D.; Orefici, I.E.; Altavilla, G.; Cusatelli, P.; Smith, I. The extracytoplasmic function sigma factor σE is essentials for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 2004, 72, 3038–3041. [Google Scholar] [CrossRef] [Green Version]
- Kazmierczak, M.J.; Wiedmann, M.; Boor, K.J. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 2005, 69, 527–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karls, R.; Guarner, J.; McMurray, D.N.; Birkness, K.A.; Quinn, F.D. Examination of Mycobacterium tuberculosis sigma factor mutants using low-dose aerozol infect ion of gwinea pigs suggest a role for SigC in pathogenesis. Microbiology 2006, 152, 1591–1600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMeechen, A.; Roberts, M.; Cogan, T.A.; Jørgensen, F.; Stevenson, A.; Lewis, C.; Rowley, G.; Humphrey, T.J. Role of the alternative sigma factors σE and σS in survival of Salmonella enterica serovar Typhimurium during starvation, refrigeration and osmotic shock. Microbiology 2007, 15, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Otero-Asman, J.R.; Quesada, J.M.; Jim, K.K.; Ocampo-Sosa, A.; Civantos, C.; Bitter, W.; Llamas, M.A. The extracytoplasmic function sigma factor σVrel is active during infection and contributes to phosphate starvation-induced virulence of Pseudomonas aeruginosa. Sci. Rep. 2020, 10, 3139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kędzierska-Mieszkowska, S.; Potrykus, K.; Arent, Z.; Krajewska, J. Identification of σE-dependent promoter upstream of clpB from the pathogenic spirochaete Leptospira interrogans by applying an E. coli two-plasmid system. Int. J. Mol. Sci. 2019, 20, 6325. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. World Health Organization. 2003. Available online: https://apps.who.int/iris/handle/10665/42667 (accessed on 17 February 2023).
- Wolff, J.W. The Laboratory Diagnosis of Leptospirosis; Publ. No. 183, American Lecture Series; Charles, C., Ed.; Thomas: Springfield, IL, USA, 1954. [Google Scholar]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Krajewska, J.; Arent, Z.; Zolkiewski, M.; Kędzierska-Mieszkowska, S. Immunoreactivity of the AAA+ chaperone ClpB from Leptospira interrogans with sera from Leptospira-infected animals. BMC Microbiol. 2016, 16, 151. [Google Scholar] [CrossRef] [Green Version]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Fjgurnov, M. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Casas-Pastor, D.; Müller, R.R.; Jaenicke, S.; Brinkrolf, K.; Becker, A.; Buttner, M.J.; Gross, C.A.; Mascher, T.; Goesmann, A.; Fritz, G. Expansion and re-classification of the extracytoplasmic function (ECF) σ factor family. Nucleic Acids Res. 2021, 49, 986–1005. [Google Scholar] [CrossRef]
- Mecsas, J.; Rouvière, P.E.; Erickson, J.W.; Donohue, T.J.; Gross, C.A. The activity of σE, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993, 7, 2618–2628. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.; Everard, C.O.R.; Faine, S.; Adler, B. Antigens recognized by the human immune response to serve leptospirosis in Barbados. Epidemiol. Infect. 1991, 107, 143–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerreiro, H.; Croda, J.; Flannery, B.; Mazel, M.; Matsunaga, J.; Reis, M.G.; Levett, P.N.; Ko, A.I.; Haake, D.A. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 2001, 69, 4958–4968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafetá, B.N.; de Castro, E.C.; da Silva, N. Protein and antigen profiles of Leptospira interrogans serovar Hardjo. Cien. Rural. 2009, 39, 2539–2543. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.A.; Lefebvre, R.; Pan, M.J. Protein and antigen profiles of prevalent serovars of Leptospira interrogans. Infect. Immun. 1991, 59, 1772–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Factor | Controlled Genes/Proposed Function |
|---|---|
| σ70 (primary σ factor) | mainly the housekeeping genes (>1000 genes)—most genes expressed during exponential growth |
| σ28 | genes encoding components of the endoflagellum (flaA1, flaB1, flaB4) and the flagellin-specific chaperone FliS (fliS); mainly cell motility |
| 11 ECF σE-type factors | extracytoplasmic function (469 putative binding sites in the L. interrogans genome for all ECF σs); heat shock response, stress survival and virulence (clpB) |
| σ54 | genes encoding putative lipoprotein and the ammonium transporter AmtB; mainly control of internal concentration of ammonia and environmental adaptation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kędzierska-Mieszkowska, S.; Arent, Z. Immunoreactivity of a Putative ECF σ Factor, LIC_10559, from Leptospira interrogans with Sera from Leptospira-Infected Animals. Pathogens 2023, 12, 512. https://doi.org/10.3390/pathogens12040512
Kędzierska-Mieszkowska S, Arent Z. Immunoreactivity of a Putative ECF σ Factor, LIC_10559, from Leptospira interrogans with Sera from Leptospira-Infected Animals. Pathogens. 2023; 12(4):512. https://doi.org/10.3390/pathogens12040512
Chicago/Turabian StyleKędzierska-Mieszkowska, Sabina, and Zbigniew Arent. 2023. "Immunoreactivity of a Putative ECF σ Factor, LIC_10559, from Leptospira interrogans with Sera from Leptospira-Infected Animals" Pathogens 12, no. 4: 512. https://doi.org/10.3390/pathogens12040512
APA StyleKędzierska-Mieszkowska, S., & Arent, Z. (2023). Immunoreactivity of a Putative ECF σ Factor, LIC_10559, from Leptospira interrogans with Sera from Leptospira-Infected Animals. Pathogens, 12(4), 512. https://doi.org/10.3390/pathogens12040512









