Novel Identification of the Collection of Pathogenic Fungal Species Verticillium with the Development of Species-Specific SSR Markers
Abstract
:1. Introduction
2. Materials and Methods
3. Results
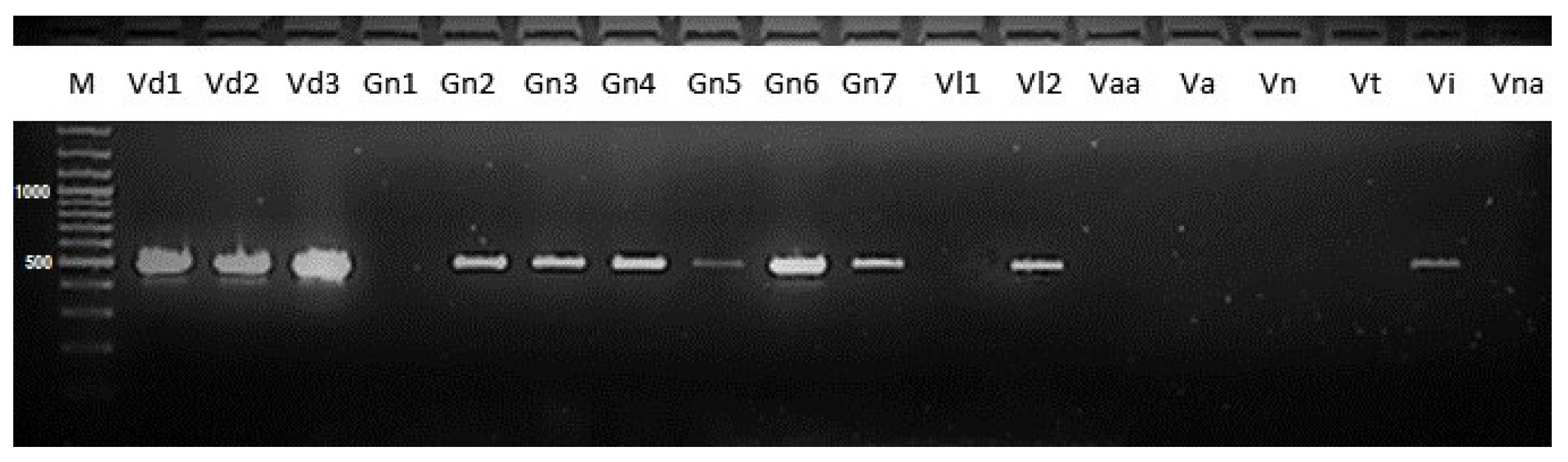
3.1. Simplex and Multiplex PCR Assays
3.2. Phylogenetic Analysis Based on ITS Sequencing
3.3. Specificity of Verticillium dahliae Simplex PCR Assay
3.4. SSR Marker Development
3.5. LAMP Sensitivity for Species Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Isolate Designation | Host or Substrate | Geographical Origin | Pathogenicity Group | Source | Morphological Identification | Identification with Simplex or Multiplex PCR Assay | Identification with NCBI Database | Identification with Phylogenetic Analysis | Identification with SSR Phylogenetic Analysis | Used in LAMP Reaction |
|---|---|---|---|---|---|---|---|---|---|---|
| CBS 102.464 | artichoke | Italy | n.t. | CBS | V. albo-atrum | V. alboatrum | V. albo-atrum strain VICVaa1 | V. albo-atrum | not amplified | No |
| CBS 682.88 | potato | Netherland | n.t. | CBS | V. albo-atrum | V. alboatrum | V. albo-atrum | V. albo-atrum | not included | No |
| 110 | potato | Prince Edwards Islands, Canada | n.t. | 13 | V. albo-atrum | V. alboatrum | V. albo-atrum | V. albo-atrum | not included | Yes |
| PD693 | tomato | Great Britain | n.t. | 12 | V. albo-atrum | V. alboatrum | V. albo-atrum | V. albo-atrum | not amplified | No |
| 166 | potato | Netherlands | n.t. | 6 | V. albo-atrum | V. albo-atrum | V. albo-atrum | V. albo-atrum | not included | No |
| 112 | potato | Prince Edwards Islands, Canada | n.t. | 6 | V. albo-atrum | V. albo-atrum | V. albo-atrum | V. albo-atrum | not included | Yes |
| Luc | alfalfa | Great Britain | lethal | 7 | V. albo-atrum | V. alfalfae | V. albo-atrum | V. alfalfae | V. alfalfae | No |
| 41 | alfalfa | Canada | n.t. | 6 | V. albo-atrum | V. alfalfae | V. albo-atrum | V. alfalfae | V. alfalfae | No |
| CBS 392.91 | alfalfa | Netherland | n.t. | CBS | V. albo-atrum | V. alfalfae | V. albo-atrum | V. alfalfae | V. alfalfae | No |
| 107 | alfalfa | ZDA | n.t. | 6 | V. albo-atrum | V. alfalfae | V. albo-atrum | V. alfalfae | V. alfalfae | Yes |
| 11 | alfalfa | Canada | n.t. | 6 | V. albo-atrum | V. alfalfae | V. albo-atrum | V. alfalfae | V. alfalfae | Yes |
| CIG3 | hop | Slovenia | mild | IHPS | V. albo-atrum | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| JKG 2 | catalpa | Netherland | n.t. | 8 | V. dahliae | V. dahliae | V. longisporum/V. dahliae | V. dahliae | V. dahliae | No |
| JKG1 | potato | Netherland | n.t. | 8 | V. dahliae | V. dahliae | V. longisporum/V. dahliae | V. dahliae | V. dahliae | No |
| JKG 8 | potato | Netherland | n.t. | 8 | V. dahliae | V. dahliae | V. longisporum/V. dahliae | V. dahliae | V. dahliae | No |
| DJK | chrysanthemum | Netherland | n.t. | 8 | V. dahliae | V. dahliae | V. dahliae | V. dahliae | V. dahliae | No |
| MAI | chrysanthemum | Netherland | n.t. | 8 | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| Mint | mint | ZDA | n.t. | 10 | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| GAJ09 | hop | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. dahliae | V. dahliae | V. dahliae | No |
| PDRENU | hop | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| CasD | hop | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | Yes |
| KresD | hop | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| MoD | hop | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| Oset | hop | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| 12042 | hop | Great Britain | n.t. | 1 | V. dahliae | V. dahliae | V. dahliae | V. dahliae | V. dahliae | No |
| PAPmb | pepper | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | G. nigrescens strain BM-1 | G. nigrescens | not amplified | No |
| PAP | pepper | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| Pap99 | pepper | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. dahliae | V. dahliae | V. dahliae | No |
| Pap2008 | pepper | Slovenia | n.t. | IHPS | V. dahliae | V. dahliae | V. dahliae | V. dahliae | V. dahliae | Yes |
| 14 | olive | Greece | VCG2 | 13 | V. dahliae | V. dahliae | V. dahliae strain Le1344 | V. dahliae | not included | No |
| 141 | olive | Bari, Italy | tomato race2 | 13 | V. dahliae | V. dahliae | V. dahliae | V. dahliae | not included | No |
| 3V | olive | Greece | VCG4 | 13 | V. dahliae | V. dahliae | V. longisporum | V. dahliae | not included | No |
| 802-1 | olive | Crete, Greece | VCG4, tomato race2 | 13 | V. dahliae | V. dahliae | V. albo-atrum | No quality sequence | not included | No |
| V 138 I | cotton | Upper Guadalquivir valley, Spain | lethal | 11 | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| V 176 I | cotton | Upper Guadalquivir valley, Spain | mild | 11 | V. dahliae | V. dahliae | V. dahliae strain Le1344 | V. dahliae | V. dahliae | No |
| PD335 | mint | ZDA | n.t. | 12 | V. dahliae | V. dahliae | V. dahliae | V. dahliae | V. dahliae | No |
| PD584 | cabbage | Japan | n.t. | 12 | V. dahliae | V. dahliae | V. longisporum | V. dahliae | V. dahliae | No |
| A III 25 | cauliflower | ZDA | n.t. | 10 | V. dahliae | V. dahliae | V. dahliae | V. dahliae | not included | No |
| PD337 | cotton | ZDA | n.t. | 12 | V. dahliae | V. dahliae | V. dahliae | V. dahliae | not included | No |
| JKG 20 | linden | Netherland | n.t. | 8 | V. tricorpus | V. isaacii | V. tricorpus | V. isaacii | not amplified | No |
| 115 | potato | Netherland | n.t. | 6 | V. albo-atrum | V. isaacii | V. tricorpus | V. isaacii | not included | No |
| EX5 F8 | soil | Netherland | n.t. | 8 | V. tricorpus | V. isaacii | V. tricorpus | V. isaacii | not included | No |
| CBS110218 | brassica napus | Sweden | n.t. | CBS | V. longisporum | V. longisporum A1/D1 | V. dahliae var. longisporum | V. longisporum | V. longisporum | No |
| PD330 | cabbage | ZDA | n.t. | 12 | V. longisporum | V. longisporum A1/D1 | V. longisporum | V. longisporum | V. longisporum | No |
| P10 | hop | Germany | lethal | 5 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| P114/1 | hop | Germany | lethal | 5 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| P34/1 | hop | Germany | lethal | 5 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| P15 | hop | Germany | lethal | 5 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| P83 | hop | Germany | mild | 5 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| 6/99 | hop | Germany | mild | 4 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 14/93 | hop | Germany | mild | 4 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 15/98 | hop | Germany | mild | 4 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| T2 | hop | Slovenia | lethal | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| TABOR 6 | hop | Slovenia | lethal | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| Ciz/DED | hop | Slovenia | lethal | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| BIZ | hop | Slovenia | lethal | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| MO 3 | hop | Slovenia | mild | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| OCer | hop | Slovenia | mild | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| Zup | hop | Slovenia | mild | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| Rec91 | hop | Slovenia | mild | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| KRES 98 | hop | Slovenia | mild | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| 1985 | hop | Great Britain | lethal | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| 11041 | hop | Great Britain | lethal | 1 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 11055 | hop | Great Britain | lethal | 1 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 11047 | hop | Great Britain | lethal | 1 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | No quality sequence | V. nonalfalfae | No |
| 11100 | hop | Great Britain | lethal | 9 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | Yes |
| 1974 | hop | Great Britain | lethal | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 298101 | hop | Great Britain | lethal | CABI | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 298102 | hop | Great Britain | lethal | CABI | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 1953 | hop | Great Britain | mild | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 298092 | hop | Great Britain | mild | CABI | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 298095 | hop | Great Britain | mild | CABI | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| Sol | hop | Poland | mild | 3 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| Surf | petunia | Slovenia | n.t. | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 11081 | chrysanthemum | Great Britain | n.t. | 1 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 11066 | potato | Great Britain | n.t. | 1 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| T 179 | tomato | Great Britain | mild | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| CBS 321.91 | tomato | Netherland | n.t. | CBS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| AR01/067 | tomato | Great Britain | lethal | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| AR0/140 | tomato | Great Britain | lethal | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| AR01/JS1 | tomato | Great Britain | n.t. | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| PD 83/53a | tomato | Netherland | n.t. | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| PD 2000/4186a | tomato | Netherland | n.t. | 2 | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 314193 | potato | Australia | n.t. | CABI | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| SN 10 | hop | Slovenia | lethal | IHPS | V. albo-atrum | V. nonalfalfae | V. albo-atrum | V. nonalfalfae | not included | No |
| CBS 456.51 | potato | Great Britain | n.t. | CBS | V. nubilum | V. nubilum | V. albo-atrum | V. nubilum | not amplified | No |
| CBS 227.84 | potato | Netherland | n.t. | CBS | V. tricorpus | V. tricorpus | V. tricorpus | V. tricorpus | not included | No |
| EX5 F7 | soil | Netherland | n.t. | 8 | V. tricorpus | V. tricorpus | V. tricorpus | V. tricorpus | not included | No |
| PETROL | hop | Slovenija | lethal | IHPS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | not included | No |
| P84/2 | hop | Germany | mild | 5 | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| KV11 | hop | Slovenia | lethal | IHPS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | not included | No |
| VranBis09 | hop | Slovenia | lethal | IHPS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| Sent4 | hop | Slovenia | lethal | IHPS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 11097 | hop | Great Britain | lethal | 1 | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 298100 | hop | Great Britain | lethal | CABI | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| CBS 393.91 | hop | Belgium | mild | CBS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| Kum | cucumber | Slovenia | n.t. | IHPS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 11077 | Galinsoga | Great Britain | n.t. | 1 | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| CBS 454.51 | potato | Great Britain | n.t. | CBS | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| 340646 | potato | Spain | n.t. | CABI | V. albo-atrum | / | V. albo-atrum | V. nonalfalfae | V. nonalfalfae | No |
| CBS 123.176 | insulation wool | Finland | n.t. | CBS | V. nigrescens | / | Unknown Verticillium | G. nigrescens | not amplified | No |
| CBS 171.80 | Agaricus bisporus | Netherland | n.t. | CBS | V. fungicola | / | V. fungicola | / | V. fungicola | No |
| CBS 122.175 | Cave cricket | Spain | n.t. | CBS | V. lecanii | / | V. leicanii | / | not amplified | No |
| B 560 | Cave cricket | Slovenia | n.t. | IHPS | V. lecanii | / | V. leicanii | / | not amplified | No |
References
- Inderbitzin, P.; Davis, R.M.; Bostock, R.M.; Subbarao, K.V. Identification and Differentiation of Verticillium Species and V. longisporum Lineages by Simplex and Multiplex PCR Assays. PLoS ONE 2013, 8, e65990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inderbitzin, P.; Bostock, R.M.; Davis, R.M.; Usami, T.; Platt, H.W.; Subbarao, K.V. Phylogenetics and Taxonomy of the Fungal Vascular Wilt Pathogen Verticillium, with the Descriptions of Five New Species. PLoS ONE 2011, 6, e28341. [Google Scholar] [CrossRef] [PubMed]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, Pathogenicity, and Management of Verticillium Species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennypacker, B.W. Verticillium wilts. Q. Rev. Biol. 2004, 79, 80. [Google Scholar] [CrossRef]
- Radišek, S.; Jakše, J.; Javornik, B. Development of Pathotype-Specific SCAR Markers for Detection of Verticillium albo-atrum Isolates from Hop. Plant Dis. 2004, 88, 1115–1122. [Google Scholar] [CrossRef] [Green Version]
- Radišek, S.; Jakše, J.; Javornik, B. Genetic variability and virulence among Verticillium albo-atrum isolates from hop. Eur. J. Plant Pathol. 2006, 116, 301–314. [Google Scholar] [CrossRef]
- Zare, R.; Gams, W.; Starink-Willemse, M.; Summerbell, R.C. Gibellulopsis, a suitable genus for Verticillium nigrescens, and Musicillium, a new genus for V. theobromae. Nova Hedwig. 2007, 85, 463–489. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk-Fradin, E.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Smith, H.C. The morphology of Verticillium albo-atrum, V. dahliae, and V. tricorpus. N. Z. J. Agric. Res. 1965, 8, 450–478. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, S. Longevity of the verticillium wilt fungus in the laboratory and field. Phytopath 1955, 45, 180–181. [Google Scholar]
- Hu, X.-P.; Gurung, S.; Short, D.P.G.; Sandoya, G.V.; Shang, W.-J.; Hayes, R.J.; Davis, R.M.; Subbarao, K.V. Nondefoliating and Defoliating Strains from Cotton Correlate with Races 1 and 2 of Verticillium dahliae. Plant Dis. 2015, 99, 1713–1720. [Google Scholar] [CrossRef] [Green Version]
- Isaac, I. A comparative study of pathogenic isolates of Verticillium. Trans. Br. Mycol. Soc. 1949, 32, 137–157. [Google Scholar] [CrossRef]
- Isaac, I. A further comparative study of pathogenic isolates of Verticillium: V. nubilum Pethybr. and V. tricorpus sp.nov. Trans. Br. Mycol. Soc. 1953, 36, 180–195. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Pérez-Artés, E.; Jiménez-Díaz, R.M. Detection of the nondefoliating pathotype of Verticillium dahliae in infected olive plants by nested PCR. Plant Pathol. 2001, 50, 609–619. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Jurado, R.; Pérez-Artés, E.; Jiménez-Díaz, R.M. Detection of the Defoliating Pathotype of Verticillium dahliae in Infected Olive Plants by Nested PCR. Eur. J. Plant Pathol. 2002, 108, 1–13. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Parrilla-Araujo, S.; Jiménez-Díaz, R.M. Simultaneous Detection of the Defoliating and Nondefoliating Verticillium dahliae Pathotypes in Infected Olive Plants by Duplex, Nested Polymerase Chain Reaction. Plant Dis. 2003, 87, 1487–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inderbitzin, P.; Subbarao, K.V. Verticillium Systematics and Evolution: How Confusion Impedes Verticillium Wilt Management and How to Resolve It. Phytopathology 2014, 104, 564–574. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, K.; Komatsu, T.; Suga, H. Refined PCR protocol for detection of plant pathogens in soil. J. Gen. Plant Pathol. 2003, 69, 153–160. [Google Scholar] [CrossRef]
- Jecz, P.; Jecz, T.; Korbin, M. The suitability of PCR-based techniques for detecting verticillium dahliae in strawberry plants and soil. J. Fruit Ornam. Plant Res. 2018, 16, 295–304. [Google Scholar]
- Maruthachalam, K.; Atallah, Z.K.; Vallad, G.E.; Klosterman, S.J.; Hayes, R.J.; Davis, R.M.; Subbarao, K.V.; Radionenko, M.; Hanson, S.F.; Sandoya, G.V.; et al. Molecular Variation Among Isolates of Verticillium dahliae and Polymerase Chain Reaction-Based Differentiation of Races. Phytopathology 2010, 100, 1222–1230. [Google Scholar] [CrossRef] [Green Version]
- Platt, H.W.; Mahuku, G. Detection methods forVerticillium species in naturally infested and inoculated soils. Am. J. Potato Res. 2000, 77, 271–274. [Google Scholar] [CrossRef]
- Radišek, S.; Jakše, J.; Simončič, A.; Javornik, B. Characterization of Verticillium albo-atrum Field Isolates Using Pathogenicity Data and AFLP Analysis. Plant Dis. 2003, 87, 633–638. [Google Scholar] [CrossRef] [Green Version]
- Robb, J.; Moukhamedov, R.; Hu, X.; Platt, H.; Nazar, R. Putative subgroups of Verticillium albo-atrum distinguishable by PCR-based assays. Physiol. Mol. Plant Pathol. 1993, 43, 423–436. [Google Scholar] [CrossRef]
- Tran, V.T.; Braus-Stromeyer, S.A.; Timpner, C.; Braus, G.H. Molecular diagnosis to discriminate pathogen and apathogen species of the hybrid Verticillium longisporum on the oilseed crop Brassica napus. Appl. Microbiol. Biotechnol. 2013, 97, 4467–4483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazar, R.; Hu, X.; Schmidt, J.; Culham, D.; Robb, J. Potential use of PCR-amplified ribosomal intergenic sequences in the detection and differentiation of verticillium wilt pathogens. Physiol. Mol. Plant Pathol. 1991, 39, 1–11. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [Green Version]
- Berbegal, M.; Garzón, C.D.; Ortega, A.; Armengol, J.; Jiménez-Díaz, R.M.; Jiménez-Gasco, M.M. Development and application of new molecular markers for analysis of genetic diversity in Verticillium dahliae populations. Plant Pathol. 2011, 60, 866–877. [Google Scholar] [CrossRef]
- Barbara, D.J.; Morton, A.; Miller, N. Isolation of microsatellite markers from an interspecific hybrid isolate of the fungal plant pathogen Verticillium dahliae. Mol. Ecol. Notes 2005, 5, 854–856. [Google Scholar] [CrossRef]
- Sampaio, P.; Gusmão, L.; Alves, C.; Pina-Vaz, C.; Amorim, A.; Pais, C. Highly Polymorphic Microsatellite for Identification of Candida albicans Strains. J. Clin. Microbiol. 2003, 41, 552–557. [Google Scholar] [CrossRef] [Green Version]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogovšek, P.; Hodgetts, J.; Hall, J.; Prezelj, N.; Nikolić, P.; Mehle, N.; Lenarčič, R.; Rotter, A.; Dickinson, M.; Boonham, N.; et al. LAMP assay and rapid sample preparation method for on-site detection of flavescence dorée phytoplasma in grapevine. Plant Pathol. 2015, 64, 286–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, X. Reverse Transcription Loop-Mediated Isothermal Amplification of DNA for Detection of Potato virus Y. Plant Dis. 2005, 89, 605–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niessen, L.; Luo, J.; Denschlag, C.; Vogel, R.F. The application of loop-mediated isothermal amplification (LAMP) in food testing for bacterial pathogens and fungal contaminants. Food Microbiol. 2013, 36, 191–206. [Google Scholar] [CrossRef]
- Bühlmann, A.; Pothier, J.F.; Rezzonico, F.; Smits, T.H.; Andreou, M.; Boonham, N.; Duffy, B.; Frey, J.E. Erwinia amylovora loop-mediated isothermal amplification (LAMP) assay for rapid pathogen detection and on-site diagnosis of fire blight. J. Microbiol. Methods 2013, 92, 332–339. [Google Scholar] [CrossRef]
- Tomlinson, J.A.; Dickinson, M.J.; Boonham, N. Rapid Detection of Phytophthora ramorum and P. kernoviae by Two-Minute DNA Extraction Followed by Isothermal Amplification and Amplicon Detection by Generic Lateral Flow Device. Phytopathology 2010, 100, 143–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moradi, A.; Almasi, M.; Jafary, H.; Mercado-Blanco, J. A novel and rapid loop-mediated isothermal amplification assay for the specific detection of Verticillium dahliae. J. Appl. Microbiol. 2014, 116, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Weising, K. DNA Fingerprinting in Plants and Fungi; CRC Press: Boca Raton, FL, USA, 1995. [Google Scholar]
- Bagar, T.; Altenbach, K.; Read, N.; Benčina, M. Live-Cell Imaging and Measurement of Intracellular pH in Filamentous Fungi Using a Genetically Encoded Ratiometric Probe. Eukaryot. Cell 2009, 8, 703–712. [Google Scholar] [CrossRef] [Green Version]
- Möller, E.; Bahnweg, G.; Sandermann, H.; Geiger, H. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 1992, 20, 6115–6116. [Google Scholar] [CrossRef] [Green Version]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [Green Version]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlötterer, C. Microsatellite analyser (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakše, J.; Jelen, V.; Radišek, S.; de Jonge, R.; Mandelc, S.; Majer, A.; Curk, T.; Zupan, B.; Thomma, B.P.H.J.; Javornik, B. Genome sequence of a lethal strain of xylem-invading verticillium nonalfalfae. Genome Announc. 2018, 6, e01458-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collopy, P.; Largeteau-Mamoun, M.L.; Romaine, C.; Royse, D.J. Molecular Phylogenetic Analyses of Verticillium fungicola and Related Species Causing Dry Bubble Disease of the Cultivated Button Mushroom, Agaricus bisporus. Phytopathology 2001, 91, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gams, W.; Zaayen, A. Contribution to the taxonomy and pathogenicity of fungicolous Verticillium species. I. Taxonomy. Eur. J. Plant Pathol. 1982, 88, 57–78. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.H.J.; Chen, Z.; Henrissat, B.; Lee, Y.-H.; Park, J.; et al. Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Rubinsztein, D.C.; Amos, W.; Leggo, J.; Goodburn, S.; Jain, S.; Li, S.-H.; Margolis, R.L.; Ross, C.A.; Ferguson-Smith, M.A. Microsatellite evolution—Evidence for directionality and variation in rate between species. Nat. Genet. 1995, 10, 337–343. [Google Scholar] [CrossRef]
- Ellegren, H.; Primmer, C.R.; Sheldon, B.C. Microsatellite ‘evolution’: Directionality or bias? Nat. Genet. 1995, 11, 360–362. [Google Scholar] [CrossRef]
- Hutter, C.M.; Schug, M.D.; Aquadro, C.F. Microsatellite variation in Drosophila melanogaster and Drosophila simulans: A reciprocal test of the ascertainment bias hypothesis. Mol. Biol. Evol. 1998, 15, 1620–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marton, K.; Flajšman, M.; Radišek, S.; Košmelj, K.; Jakše, J.; Javornik, B.; Berne, S. Comprehensive analysis of Verticillium nonalfalfae in silico secretome uncovers putative effector proteins expressed during hop invasion. PLoS ONE 2018, 13, e0198971. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Branch Designation | Isolates Forming Groups with the Same ITS Sequence |
|---|---|
| V. alboatrum 5 | 110, 112, 166, PD693, CBS 682.88 |
| V. alboatrum artichoke 1 | CBS 102.464 |
| V. isaacii 3 | 115, EX5 F8, JKG 20 |
| V. tricorpus 2 | EX5 F7, CBS 227.84 |
| V. dahliae 2 cotton olive | 14, V-176 I |
| V. dahliae 24 | AIII25, PD337, JKG2, Mint, KresD, CasD, CIG3, 3V, V-138 I, Pap99, Pap2008, PDRENU, PD584, PD335, PAP, Oset, MoD, MAI, JKG8, JKG1, GAJ09, DJK, 141, 12042 |
| V. nonalfalfae 53 | 340646, 314139, 298101, 298102, 298100, 298092, 1985a, 15/99, 11097, 11081, 11077, 11066, 11055, 11041, P10, P114/1, KRES98, 1953, 14/93, KV11, Zup, VranBis09, T-179, TABOR6, T2, Surf, Sol, Sent04, Rec91, PETROL, PD83/53a, P84/2, P34/1, P15, OCer, MO 3, PD2000/4186a, Kum, Ciz, CBS 454.51, CBS 393.91, CBS 321.91, BIZ, AR0/140, AR01/JS1, AR01/067 |
| V. longisporum A1_D1 2 | PD330, CBS 110.218 |
| V. alfalfae 5 | Luc, Kanada11, 107, 41, CBS 392.91 |
| V. nubilum 1 | CBS 456.51 |
| G. nigrescens 1 | PapMB |
| G. nigrescens 1 | CBS 123.176 |
| SSR Locus Designation | Motif | Length (bp) | Forward Primer 5′-3′ | Reverse Primer 5′-3′ |
|---|---|---|---|---|
| 598 | (AG)16 | 177 | TGTGAGGGCACTGACATGAT | AGAACAGCCTCTTCCGTCAA |
| 959 | (GA)19 | 213 | CCAACCCTTCCATCACATTT | TGGAGGCAGTGATGAGTTTG |
| 228 | (AGA)14 | 200 | CCGGACGAGAGAGTCTGAAG | TTTGAGCAGATTATGCGTCG |
| 104 | (CT)21 | 188 | CCCTTTTCCCATCTTCAACA | TGACCGAGGAGGAGTTTGAC |
| 886 | (TG)25 | 160 | CAGCAAGGAAGCACTGTCAA | TCCAGACTACACTCTCGCCC |
| 2756 | (GT)17 | 226 | TACGATGCGCTCTCAGAATG | CTCCTGTCAGAAGGTCCAGC |
| 3507 | (GA)17 | 197 | AGAGGATGGCATGTCTGGAT | AGACAGTCTGCCTTGCCAAT |
| 1468 | (TCT)10 | 171 | GATGCGGTTCTTTGTGGACT | CAAAGGGTCATGGTGTCAGA |
| 2390 | (CT)17 | 145 | CCATGGTCGTATCTCGTGTG | GGCGGCTCTACTTCAATCAG |
| 3111 | (GA)24 | 200 | TGTAAGCTTTGCGTGACCTG | CCCTGAGCCAATTTATCGAA |
| 1566 | (CT)16 | 130 | TCGGATCCCAGGAGTAAGTTT | GCACGAGCTGGAAATTCTGT |
| 3632 | (GA)15 | 230 | GACGTGGAGAAGGTGGAGAG | GCCGCCTATGTAAGCAAAGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeseničnik, T.; Kaurin, A.; Grgič, Z.; Radišek, S.; Jakše, J.; Štajner, N. Novel Identification of the Collection of Pathogenic Fungal Species Verticillium with the Development of Species-Specific SSR Markers. Pathogens 2023, 12, 535. https://doi.org/10.3390/pathogens12040535
Jeseničnik T, Kaurin A, Grgič Z, Radišek S, Jakše J, Štajner N. Novel Identification of the Collection of Pathogenic Fungal Species Verticillium with the Development of Species-Specific SSR Markers. Pathogens. 2023; 12(4):535. https://doi.org/10.3390/pathogens12040535
Chicago/Turabian StyleJeseničnik, Taja, Anela Kaurin, Zarja Grgič, Sebastjan Radišek, Jernej Jakše, and Nataša Štajner. 2023. "Novel Identification of the Collection of Pathogenic Fungal Species Verticillium with the Development of Species-Specific SSR Markers" Pathogens 12, no. 4: 535. https://doi.org/10.3390/pathogens12040535
APA StyleJeseničnik, T., Kaurin, A., Grgič, Z., Radišek, S., Jakše, J., & Štajner, N. (2023). Novel Identification of the Collection of Pathogenic Fungal Species Verticillium with the Development of Species-Specific SSR Markers. Pathogens, 12(4), 535. https://doi.org/10.3390/pathogens12040535













