NRF2 Antioxidant Response and Interferon-Stimulated Genes Are Differentially Expressed in Respiratory-Syncytial-Virus- and Rhinovirus-Infected Hospitalized Children
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Gene Expression Measurement
2.3. Statistical Analysis
3. Results
3.1. RSV and HRV Detection and Clinical Diagnosis in the Study Groups
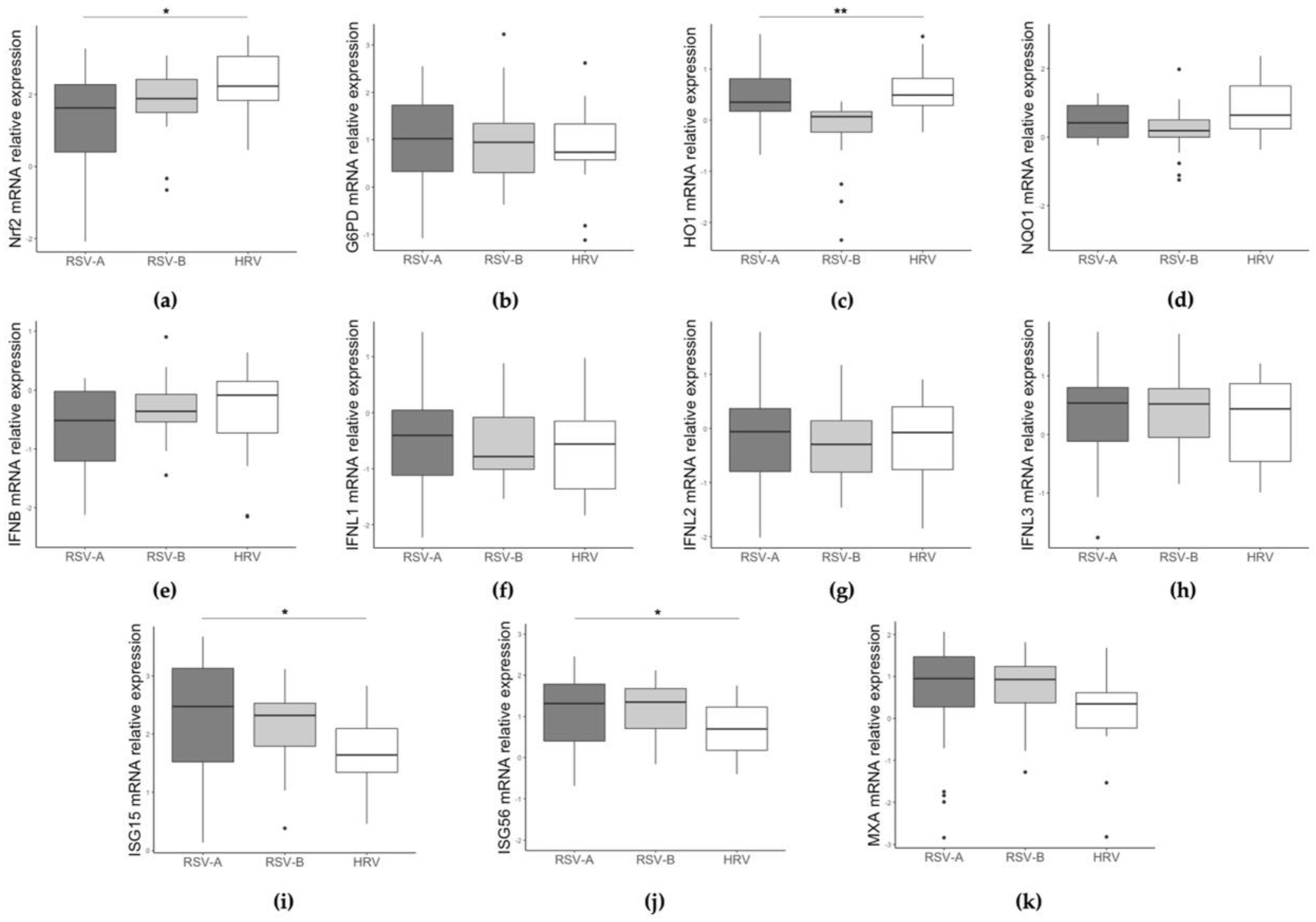
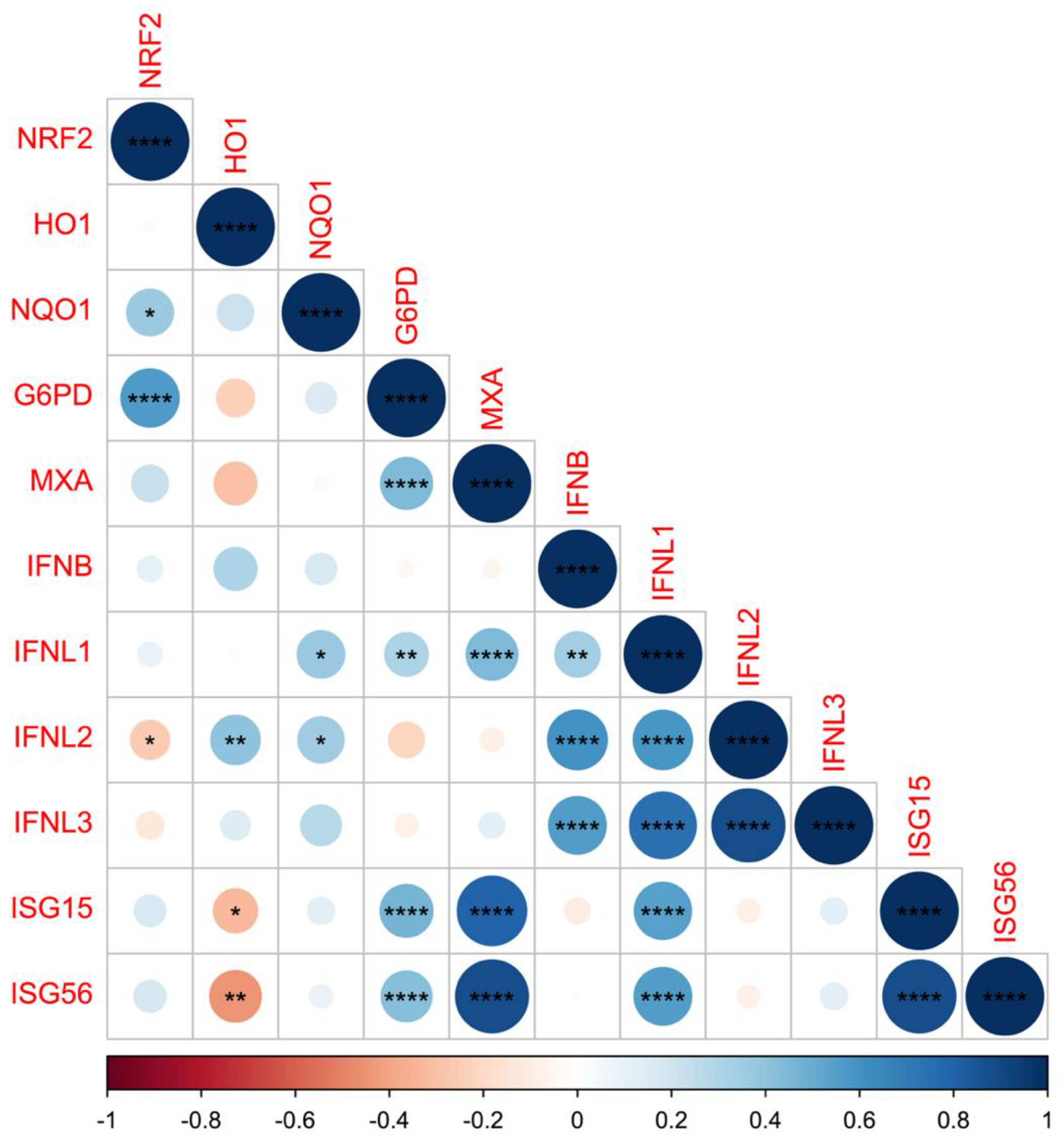
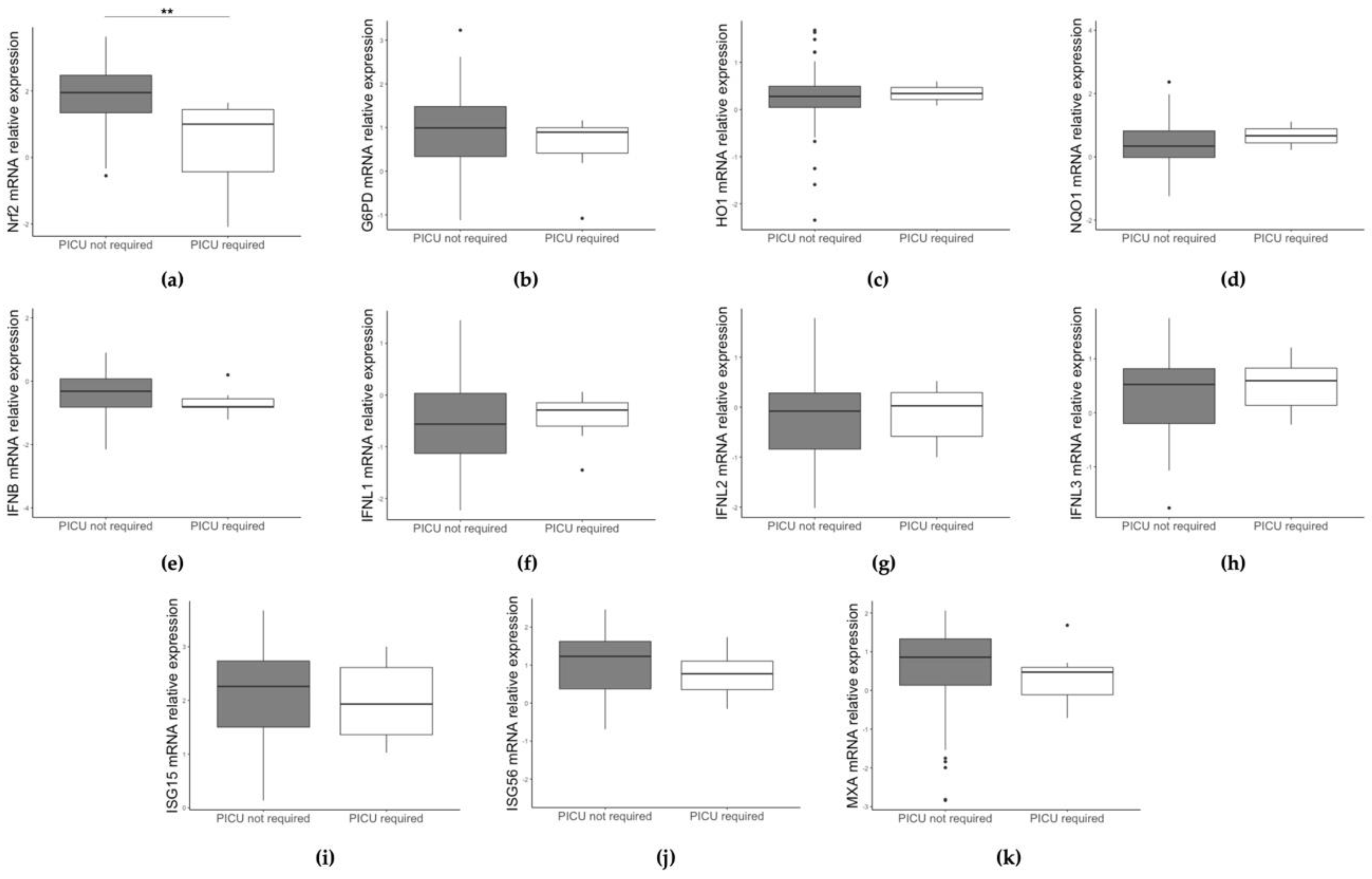
3.2. Different Gene Expression Based on RSV/HRV Status and Clinical Data
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nair, H.; Nokes, D.J.; Gessner, B.D.; Dherani, M.; Madhi, S.A.; Singleton, R.J.; O’Brien, K.L.; Roca, A.; Wright, F.P.; Bruce, N.; et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet 2010, 375, 1545–1555. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, X.; Blau, D.M.; Caballero, T.M.; Feikin, D.R.; Gill, C.J.; Madhi, S.A.; Omer, S.B.; Simoes, E.A.; Campbell, H.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: A systematic analysis. Lancet 2022, 399, 2047–2064. [Google Scholar] [CrossRef]
- Smyth, R.L.; Openshaw, P.J.M. Bronchiolitis. Lancet 2006, 368, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Hennessey, P.A.; Formica, M.A.; Cox, C.; Walsh, E.E. Respiratory syncytial virus infection in elderly and high-risk adults. N. Engl. J. Med. 2005, 28, 1749–1759. [Google Scholar] [CrossRef]
- Meissner, H.C. Viral Bronchiolitis in Children. N. Engl. J. Med. 2016, 374, 62–72. [Google Scholar] [CrossRef]
- Bouzid, D.; Hadad, O.; Bertine, M.; Houhou-Fidouh, N.; Mirand, A.; Duval, X.; Bunel, V.; Borie, R.; Lucet, J.C.; Descamps, D.; et al. Rhinoviruses: Molecular diversity and clinical characteristics. Int. J. Infect. Dis. 2022, 1201-9712, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Calışkan, M.; Bochkov, Y.A.; Kreiner-Møller, E.; Bønnelykke, K.; Stein, M.M.; Du, G.; Bisgaard, H.; Jackson, D.J.; Gern, J.E.; Lemanske, R.F.; et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N. Engl. J. Med. 2013, 368, 1398–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drysdale, S.B.; Mejias, A.; Ramilo, O. Rhinovirus—Not just the common cold. J. Infect. 2017, 74, S41–S46. [Google Scholar] [CrossRef] [PubMed]
- Midulla, F.; Scagnolari, C.; Bonci, E.; Pierangeli, A.; Antonelli, G.; De Angelis, D.; Berardi, R.; Moretti, C. Respiratory syncytial virus, human bocavirus and rhinovirus bronchiolitis in infants. Arch. Dis. Child. 2010, 95, 35–41. [Google Scholar] [CrossRef]
- Durbin, R.K.; Kotenko, S.V.; Durbin, J.E. Interferon induction and function at the mucosal surface. Immunol. Rev. 2013, 255, 25–39. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 4, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Hussell, T.; Goulding, J. Structured regulation of inflammation during respiratory viral infection. Lancet Infect. Dis. 2010, 5, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Davidson, S.; Maini, M.K.; Wack, A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J. Interferon. Cytokine Res. 2015, 35, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Do Lah Pellet, J.; van Doorn, H.R.; Tran, A.T.; Nguyen, B.H.; Tran, T.T.L.; Tran, Q.H.; Vo, Q.B.; Tran Dac, N.A.; Trinh, H.N.; Nguyen, T.T.H.; et al. Host transcription profile in nasal epithelium and whole blood of hospitalized children under 2 years of age with respiratory syncytial virus infection. J. Infect. Dis. 2017, 217, 134–146. [Google Scholar]
- Pierangeli, A.; Viscido, A.; Bitossi, C.; Frasca, F.; Gentile, M.; Oliveto, G.; Frassanito, A.; Nenna, R.; Midulla, F.; Scagnolari, C. Differential interferon gene expression in bronchiolitis caused by respiratory syncytial virus-A genotype ON1. Med. Microbiol. Immunol. 2020, 209, 23–28. [Google Scholar] [CrossRef]
- Goritzka, M.; Durant, L.R.; Pereira, C.; Salek-Ardakani, S.; Openshaw, P.J.; Johansson, C. Alpha/beta interferon receptor signaling amplifies early proinflammatory cytokine production in the lung during respiratory syncytial virus infection. J. Virol. 2014, 88, 6128–6136. [Google Scholar] [CrossRef] [Green Version]
- Fraternale, A.; Zara, C.; De Angelis, M.; Nencioni, L.; Palamara, A.T.; Retini, M.; Di Mambro, T.; Magnani, M.; Crinelli, R. Intracellular Redox-Modulated Pathways as Targets for Effective Approaches in the Treatment of Viral Infection. Int. J. Mol. Sci. 2021, 22, 3603. [Google Scholar] [CrossRef]
- De Angelis, M.; Amatore, D.; Checconi, P.; Zevini, A.; Fraternale, A.; Magnani, M.; Hiscott, J.; De Chiara, G.; Palamara, A.T.; Nencioni, L. Influenza Virus Down-Modulates G6PD Expression and Activity to Induce Oxidative Stress and Promote Its Replication. Front. Cell. Infect. Microbiol. 2022, 11, 804976. [Google Scholar] [CrossRef]
- Kim, J.; Surh, Y.J. The Role of Nrf2 in Cellular Innate Immune Response to Inflammatory Injury. Toxicol. Res. 2009, 25, 159–173. [Google Scholar] [CrossRef]
- Jaiswal, A.K. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 2004, 36, 1199–1207. [Google Scholar] [CrossRef]
- Ho, H.Y.; Cheng, M.L.; Chiu, D.T. Glucose-6-phosphate dehydrogenase—From oxidative stress to cellular functions and degenerative diseases. Redox Rep. 2007, 12, 109–118. [Google Scholar] [CrossRef]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An Overview of Nrf2 Signaling Pathway and Its Role in Inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. The diverse functionality of NQO1 and its roles in redox control. Redox Biol. 2021, 41, 101950. [Google Scholar] [CrossRef]
- Kobayashi, E.H.; Suzuki, T.; Funayama, R.; Nagashima, T.; Hayashi, M.; Sekine, H.; Tanaka, N.; Moriguchi, T.; Motohashi, H.; Nakayama, K.; et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 2016, 7, 11624. [Google Scholar] [CrossRef] [Green Version]
- Olagnier, D.; Brandtoft, A.M.; Gunderstofte, C.; Villadsen, N.L.; Krapp, C.; Thielke, A.L.; Laustsen, A.; Peri, S.; Hansen, A.L.; Bonefeld, L.; et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 2018, 9, 3506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Horst, D.; Carter-Timofte, M.E.; van Grevenynghe, J.; Laguette, N.; Dinkova-Kostova, A.T.; Olagnier, D. Regulation of innate immunity by Nrf2. Curr. Opin. Immunol. 2022, 78, 102247. [Google Scholar] [CrossRef]
- Herengt, A.; Thyrsted, J.; Holm, C.K. NRF2 in Viral Infection. Antioxidants 2021, 10, 1491. [Google Scholar] [CrossRef] [PubMed]
- Khomich, O.A.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Redox Biology of Respiratory Viral Infections. Viruses 2018, 10, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olagnier, D.; Farahani, E.; Thyrsted, J.; Blay-Cadanet, J.; Herengt, A.; Idorn, M.; Hait, A.; Hernaez, B.; Knudsen, A.; Iversen, M.B.; et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020, 11, 4938. [Google Scholar] [CrossRef]
- Cho, H.Y.; Imani, F.; Miller-Degraff, L.; Walters, D.; Melendi, G.A.; Yamamoto, M.; Polack, F.P.; Kleeberger, S.R. Antiviral activity of Nrf2 in a murine model of respiratory syncytial virus (RSV) disease. Am. J. Respir. Crit. Care Med. 2009, 179, 138–150. [Google Scholar] [CrossRef] [Green Version]
- Hosakote, Y.M.; Liu, T.; Castro, S.M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell. Mol. Biol. 2009, 41, 348–357. [Google Scholar] [CrossRef]
- Komaravelli, N.; Tian, B.; Ivanciuc, T.; Mautemps, N.; Brasier, A.R.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of Nrf2. Free Radic. Biol. Med. 2015, 88, 391–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanciuc, T.; Sbrana, E.; Casola, A.; Garofalo, R.P. Protective Role of Nuclear Factor Erythroid 2-Related Factor 2 Against Respiratory Syncytial Virus and Human Metapneumovirus Infections. Front. Immunol. 2018, 9, 854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosakote, Y.M.; Jantzi, P.D.; Esham, D.L.; Spratt, H.; Kurosky, A.; Casola, A.; Garofalo, R.P. Viral-mediated inhibition of antioxidant enzymes contributes to the pathogenesis of severe respiratory syncytial virus bronchiolitis. Am J Respir Crit Care Med. 2011, 183, 1550–1560. [Google Scholar] [CrossRef] [Green Version]
- Ansar, M.; Qu, Y.; Ivanciuc, T.; Garofalo, R.P.; Casola, A. Lack of Type I Interferon Signaling Ameliorates Respiratory Syncytial Virus-Induced Lung Inflammation and Restores Antioxidant Defenses. Antioxidants 2021, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Ganjian, H.; Rajput, C.; Elzoheiry, M.; Sajjan, U. Rhinovirus and Innate Immune Function of Airway Epithelium. Front. Cell. Infect. Microbiol. 2020, 10, 277. [Google Scholar] [CrossRef]
- Mihaylova, V.T.; Kong, Y.; Fedorova, O.; Sharma, L.; Dela Cruz, C.S.; Pyle, A.M.; Iwasaki, A.; Foxman, E.F. Regional Differences in Airway Epithelial Cells Reveal Tradeoff between Defense against Oxidative Stress and Defense against Rhinovirus. Cell Rep. 2018, 24, 3000–3007.e3. [Google Scholar] [CrossRef] [Green Version]
- Selvaggi, C.; Pierangeli, A.; Fabiani, M.; Spano, L.; Nicolai, A.; Papof, P.; Moretti, C.; Midulla, F.; Antonelli, G.; Scagnolari, C. Interferon lambda 1–3 expression in infants hospitalized for RSV or HRV associated bronchiolitis. J. Infect. 2014, 68, 467–477. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Pi, J.; Zhang, Q. Signal amplification in the KEAP1-NRF2-ARE antioxidant response pathway. Redox Biol. 2022, 54, 102389. [Google Scholar] [CrossRef]
- Han, S.; Zhuang, H.; Lee, P.Y.; Li, M.; Yang, L.; Nigrovic, P.A.; Reeves, W.H. NF-E2-Related Factor 2 Regulates Interferon Receptor Expression and Alters Macrophage Polarization in Lupus. Arthritis Rheumatol. 2020, 72, 1707–1720. [Google Scholar] [CrossRef]
- Tian, B.; Yang, J.; Zhao, Y.; Ivanciuc, T.; Sun, H.; Garofalo, R.P.; Brasier, A.R. BRD4 Couples NF-κB/RelA with Airway Inflammation and the IRF-RIG-I Amplification Loop in Respiratory Syncytial Virus Infection. J. Virol. 2017, 91, e00007-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraternale, A.; De Angelis, M.; De Santis, R.; Amatore, D.; Masini, S.; Monittola, F.; Menotta, M.; Biancucci, F.; Bartoccini, F.; Retini, M.; et al. Targeting SARS-CoV-2 by synthetic dual-acting thiol compounds that inhibit Spike/ACE2 interaction and viral protein production. FASEB J. 2023, 37, e22741. [Google Scholar] [CrossRef] [PubMed]

| Patients (N = 85) | RSV (N = 63) | HRV (N = 22) | p-Value a |
|---|---|---|---|
| Demographic and clinical data | |||
| Male/female | 36/27 | 16/6 | 0.217 |
| Age in months b | 4.91 (5.25) | 4.98 (4.68) | 0.960 |
| PICU admission | 7 (11.1%) | 0 (0.0%) | 0.182 |
| Severity score (1–3): N (%) c | 17/51 (33.3%) | 12/19 (63.2%) | 0.033 |
| Severity score (4–5): N (%) c | 18/51 (35.3%) | 6/19 (31.6%) | |
| Severity score (6–8): N (%) c | 16/51 (31.4%) | 1/19 (5.2%) | |
| Gene expression d | |||
| NRF2 | 1.77 (1.15) | 2.23 (1.22) | 0.01 |
| G6PD | 1.01 (1.14) | 0.74 (0.77) | 0.670 |
| HO1 | 0.14 (0.43) | 0.49 (0.53) | 0.007 |
| NQO1 | 0.26 (0.74) | 0.63 (1.39) | 0.354 |
| IFNBeta | −0.37 (0.85) | −0.09 (0.88) | 0.236 |
| IFNL1 | −0.49 (1.07) | −0.56 (1.21) | 0.700 |
| IFNL2 | −0.08 (1.06) | −0.08 (1.16) | 0.779 |
| IFNL3 | 0.54 (0.89) | 0.44 (1.33) | 0.627 |
| ISG15 | 2.39 (1.21) | 1.64 (0.75) | 0.006 |
| ISG56 | 1.32 (1.34) | 0.69 (1.05) | 0.014 |
| MxA | 0.93 (1.07) | 0.35 (0.85) | 0.028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorrentino, L.; Toscanelli, W.; Fracella, M.; De Angelis, M.; Frasca, F.; Scagnolari, C.; Petrarca, L.; Nenna, R.; Midulla, F.; Palamara, A.T.; et al. NRF2 Antioxidant Response and Interferon-Stimulated Genes Are Differentially Expressed in Respiratory-Syncytial-Virus- and Rhinovirus-Infected Hospitalized Children. Pathogens 2023, 12, 577. https://doi.org/10.3390/pathogens12040577
Sorrentino L, Toscanelli W, Fracella M, De Angelis M, Frasca F, Scagnolari C, Petrarca L, Nenna R, Midulla F, Palamara AT, et al. NRF2 Antioxidant Response and Interferon-Stimulated Genes Are Differentially Expressed in Respiratory-Syncytial-Virus- and Rhinovirus-Infected Hospitalized Children. Pathogens. 2023; 12(4):577. https://doi.org/10.3390/pathogens12040577
Chicago/Turabian StyleSorrentino, Leonardo, Walter Toscanelli, Matteo Fracella, Marta De Angelis, Federica Frasca, Carolina Scagnolari, Laura Petrarca, Raffaella Nenna, Fabio Midulla, Anna Teresa Palamara, and et al. 2023. "NRF2 Antioxidant Response and Interferon-Stimulated Genes Are Differentially Expressed in Respiratory-Syncytial-Virus- and Rhinovirus-Infected Hospitalized Children" Pathogens 12, no. 4: 577. https://doi.org/10.3390/pathogens12040577







