The Prevalence of Undiagnosed Salmonella enterica Serovar Typhi in Healthy School-Aged Children in Osun State, Nigeria
Abstract
:1. Introduction
2. Methodology
2.1. Study Design
2.2. Ethical Approval
2.3. Inclusion Criteria
2.4. Exclusion Criteria
2.5. Sample Collection and Processing
2.6. Antigen and Antibodies Detection Using the Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Bacterial Culture for Isolation of the Salmonella Typhi
2.8. Molecular Detection of Salmonella Typhi Using Polymerase Chain Reaction (PCR)
2.9. Whole Genome Sequencing of Salmonella Typhi Using the Next Generation Sequencing Technique
2.10. Data Analysis
2.11. Quality Control and Taxonomic Classification
3. Results
3.1. Demography of Study Participants
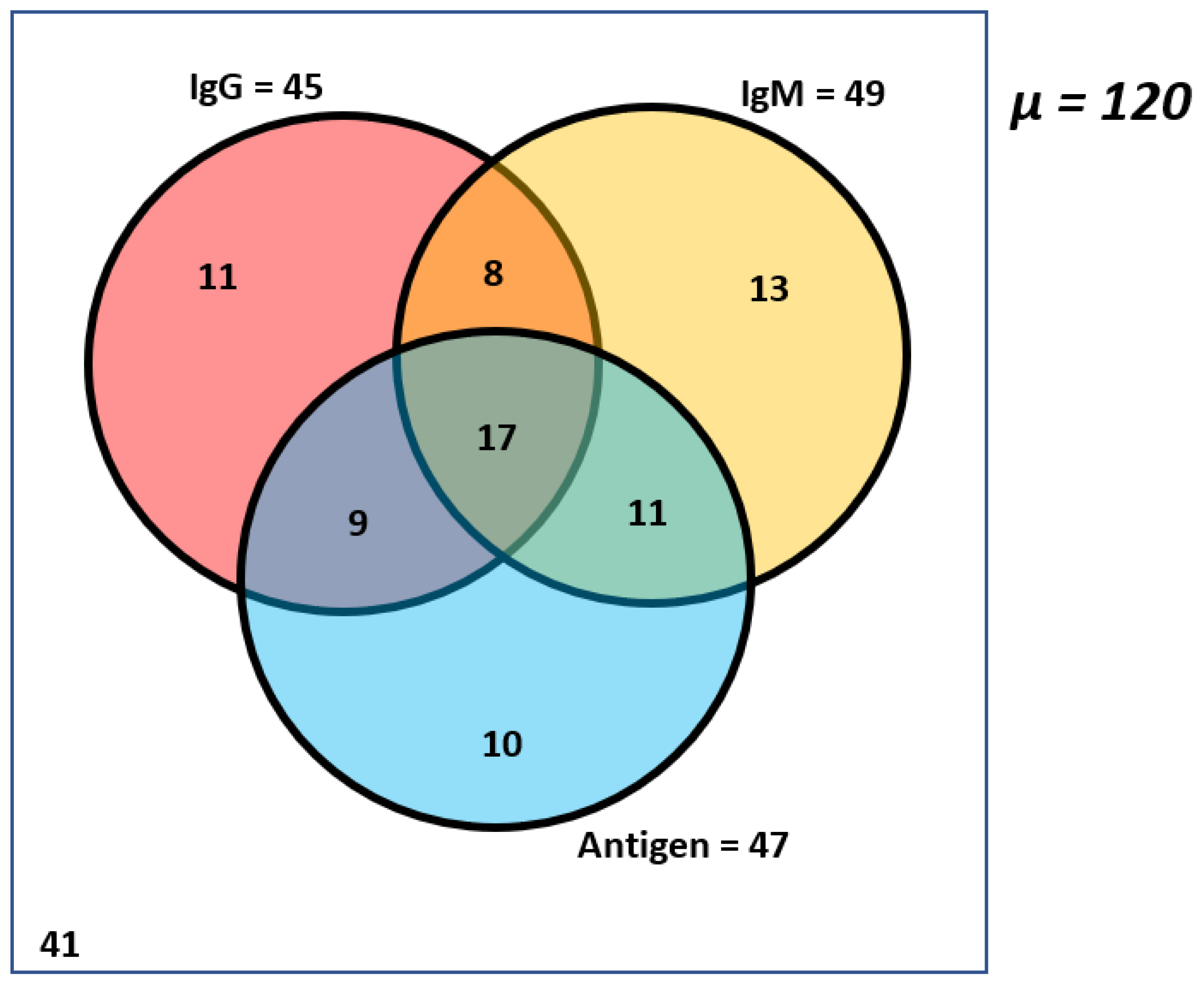
3.2. High Incidence of Salmonella Typhi Antibodies and Antigen in Non-Symptomatic Healthy Children
3.3. Bacteria Culture in Detecting Salmonella Typhi in Healthy Children
3.4. The Molecular Approach to Detecting Salmonella Typhi in Healthy Children
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Farooqui, A.; Khan, A.; Kazmi, S.U. Investigation of a Community Outbreak of Typhoid Fever Associated with Drinking Water. BMC Public Health 2009, 9, 476. [Google Scholar] [CrossRef] [Green Version]
- Bhan, M.K.; Bahl, R.; Bhatnagar, S. Typhoid and Paratyphoid Fever. Lancet 2005, 366, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Ohanu, M.E.; Iroezindu, M.O.; Maduakor, U.; Onodugo, O.D.; Gugnani, H.C. Typhoid Fever among Febrile Nigerian Patients: Prevalence, Diagnostic Performance of the Widal Test and Antibiotic Multi-Drug Resistance. Malawi Med. J. 2019, 31, 184–192. [Google Scholar] [CrossRef]
- Crump, J.A.; Luby, S.P.; Mintz, E.D. The Global Burden of Typhoid Fever. Bull. World Health Organ. 2004, 82, 346–353. [Google Scholar] [PubMed]
- Mogasale, V.; Maskery, B.; Ochiai, R.L.; Lee, J.S.; Mogasale, V.V.; Ramani, E.; Kim, Y.E.; Park, J.K.; Wierzba, T.F. Burden of Typhoid Fever in Low-Income and Middle-Income Countries: A Systematic, Literature-Based Update with Risk-Factor Adjustment. Lancet Glob. Health 2014, 2, e570–e580. [Google Scholar] [CrossRef] [Green Version]
- Antillón, M.; Warren, J.L.; Crawford, F.W.; Weinberger, D.M.; Kürüm, E.; Pak, G.D.; Marks, F.; Pitzer, V.E. The Burden of Typhoid Fever in Low- and Middle-Income Countries: A Meta-Regression Approach. PLoS Negl. Trop. Dis. 2017, 11, e0005376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-H.; Im, J.; Parajulee, P.; Holm, M.; Cruz Espinoza, L.M.; Poudyal, N.; Mogeni, O.D.; Marks, F. A Systematic Review of Typhoid Fever Occurrence in Africa. Clin. Infect. Dis. 2019, 69, S492–S498. [Google Scholar] [CrossRef]
- GBD 2017 Typhoid and Paratyphoid Collaborators. The Global Burden of Typhoid and Paratyphoid Fevers: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019, 19, 369–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akinyemi, K.O.; Oyefolu, A.O.B.; Mutiu, W.B.; Iwalokun, B.A.; Ayeni, E.S.; Ajose, S.O.; Obaro, S.K. Typhoid Fever: Tracking the Trend in Nigeria. Am. J. Trop. Med. Hyg. 2018, 99, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Wain, J.; Hendriksen, R.S.; Mikoleit, M.L.; Keddy, K.H.; Ochiai, R.L. Typhoid Fever. Lancet 2015, 385, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Neupane, D.P.; Dulal, H.P.; Song, J. Enteric Fever Diagnosis: Current Challenges and Future Directions. Pathogens 2021, 10, 410. [Google Scholar] [CrossRef]
- Gonzalez-Escobedo, G.; Marshall, J.M.; Gunn, J.S. Chronic and Acute Infection of the Gall Bladder by Salmonella Typhi: Understanding the Carrier State. Nat. Rev. Microbiol. 2011, 9, 9–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thanh Duy, P.; Thieu, N.T.V.; Nguyen Thi Nguyen, T.; Ngoc Dan Thanh, H.; Dongol, S.; Karkey, A.; Carey, M.; Basnyat, B.; Dougan, G.; Rabaa, M.A.; et al. Gallbladder Carriage Generates Genetic Variation and Genome Degradation in Salmonella Typhi. PLoS Pathog. 2020, 16, e1008998. [Google Scholar] [CrossRef] [PubMed]
- Parry, C.M.; Hien, T.T.; Dougan, G.; White, N.J.; Farrar, J.J. Typhoid Fever. N. Engl. J. Med. 2002, 347, 1770–1782. [Google Scholar] [CrossRef] [Green Version]
- Mogasale, V.V.; Ramani, E.; Mogasale, V.; Park, J.Y.; Wierzba, T.F. Estimating Typhoid Fever Risk Associated with Lack of Access to Safe Water: A Systematic Literature Review. J. Environ. Public Health 2018, 2018, 9589208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, I.; Chikwem, J.O.; Gashau, W. Determination by Widal Agglutination of the Baseline Titre for the Diagnosis of Typhoid Fever in Two Nigerian States. Scand. J. Immunol. Suppl. 1992, 11, 153–156. [Google Scholar] [CrossRef]
- Mather, R.G.; Hopkins, H.; Parry, C.M.; Dittrich, S. Redefining Typhoid Diagnosis: What Would an Improved Test Need to Look Like? BMJ Glob. Health 2019, 4, e001831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popoola, O.; Kehinde, A.; Ogunleye, V.; Adewusi, O.J.; Toy, T.; Mogeni, O.D.; Aroyewun, E.O.; Agbi, S.; Adekanmbi, O.; Adepoju, A.; et al. Bacteremia Among Febrile Patients Attending Selected Healthcare Facilities in Ibadan, Nigeria. Clin. Infect. Dis. 2019, 69, S466–S473. [Google Scholar] [CrossRef] [PubMed]
- Najib, M.A.; Mustaffa, K.M.F.; Ong, E.B.B.; Selvam, K.; Khalid, M.F.; Awang, M.S.; Zambry, N.S.; Manaf, A.A.; Bustami, Y.; Hamzah, H.H.; et al. Performance of Immunodiagnostic Tests for Typhoid Fever: A Systematic Review and Meta-Analysis. Pathogens 2021, 10, 1184. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.E.; Pan-Ngum, W.; Wijedoru, L.P.M.; Sona, S.; Nga, T.V.T.; Duy, P.T.; Vinh, P.V.; Chheng, K.; Kumar, V.; Emary, K.; et al. Evaluation of the Diagnostic Accuracy of a Typhoid IgM Flow Assay for the Diagnosis of Typhoid Fever in Cambodian Children Using a Bayesian Latent Class Model Assuming an Imperfect Gold Standard. Am. J. Trop. Med. Hyg. 2014, 90, 114–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duru, C.; Olanipekun, G.; Odili, V.; Kocmich, N.; Rezac, A.; Ajose, T.O.; Medugu, N.; Umoru, D.; Onuchukwu, C.; Munir, H.; et al. Molecular Characterization of Invasive Enterobacteriaceae from Pediatric Patients in Central and Northwestern Nigeria. PLoS ONE 2020, 15, e0230037. [Google Scholar] [CrossRef]
- House, D.; Wain, J.; Ho, V.A.; Diep, T.S.; Chinh, N.T.; Bay, P.V.; Vinh, H.; Duc, M.; Parry, C.M.; Dougan, G.; et al. Serology of Typhoid Fever in an Area of Endemicity and Its Relevance to Diagnosis. J. Clin. Microbiol. 2001, 39, 1002–1007. [Google Scholar] [CrossRef] [Green Version]
- Watson, C.H.; Baker, S.; Lau, C.L.; Rawalai, K.; Taufa, M.; Coriakula, J.; Thieu, N.T.V.; Van, T.T.; Ngoc, D.T.T.; Hens, N.; et al. A Cross-Sectional Seroepidemiological Survey of Typhoid Fever in Fiji. PLoS Negl. Trop. Dis. 2017, 11, e0005786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felgner, J.; Jain, A.; Nakajima, R.; Liang, L.; Jasinskas, A.; Gotuzzo, E.; Vinetz, J.M.; Miyajima, F.; Pirmohamed, M.; Hassan-Hanga, F.; et al. Development of ELISAs for Diagnosis of Acute Typhoid Fever in Nigerian Children. PLoS Negl. Trop. Dis. 2017, 11, e0005679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, H.; Diallo, S.; Tennant, S.M.; Livio, S.; Sow, S.O.; Tapia, M.; Fields, P.I.; Mikoleit, M.; Tamboura, B.; Kotloff, K.L.; et al. PCR Method to Identify Salmonella Enterica Serovars Typhi, Paratyphi A, and Paratyphi B among Salmonella Isolates from the Blood of Patients with Clinical Enteric Fever. J. Clin. Microbiol. 2008, 46, 1861–1866. [Google Scholar] [CrossRef] [Green Version]
- Gernert, K.M.; Seby, S.; Schmerer, M.W.; Thomas, J.C.; Pham, C.D.; St Cyr, S.; Schlanger, K.; Weinstock, H.; Shafer, W.M.; Raphael, B.H.; et al. Azithromycin Susceptibility of Neisseria Gonorrhoeae in the USA in 2017: A Genomic Analysis of Surveillance Data. Lancet Microbe 2020, 1, e154–e164. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Park, D.; Tomkins-Tinch, C.; Ye, S.; Jungreis, I.; Shlyakhter, I.; Metsky, H.; Hanna; Krasilnikova, L.A.; Lin, M.; Lin, A.; et al. Broadinstitute/Viral-Pipelines: v2.1.18.0. 2021. Available online: https://github.com/broadinstitute/viral-ngs (accessed on 1 November 2022).
- Khanam, F.; Sayeed, M.A.; Choudhury, F.K.; Sheikh, A.; Ahmed, D.; Goswami, D.; Hossain, M.L.; Brooks, A.; Calderwood, S.B.; Charles, R.C.; et al. Typhoid Fever in Young Children in Bangladesh: Clinical Findings, Antibiotic Susceptibility Pattern and Immune Responses. PLoS Negl. Trop. Dis. 2015, 9, e0003619. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.H.; Jain, A.; Nakajima, R.; Liang, L.; Jasinskis, A.; Supnet, M.; Felgner, P.L.; Teng, A.; Pablo, J.; Molina, D.M.; et al. Serodiagnosis of Acute Typhoid Fever in Nigerian Pediatric Cases by Detection of Serum IgA and IgG Against Hemolysin E and Lipopolysaccharide. Am. J. Trop. Med. Hyg. 2016, 95, 431–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijedoru, L.; Mallett, S.; Parry, C.M. Rapid Diagnostic Tests for Typhoid and Paratyphoid (enteric) Fever. Cochrane Database Syst. Rev. 2017, 5, CD008892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, B.; Rongsen-Chandola, T.; Goyal, N.; Arya, A.; Kumar, C.M.; Chakravarty, A.; Aslam, M.; More, D.; SEFI Tier 1 Collaborators. Incidence of Enteric Fever in a Pediatric Cohort in North India: Comparison with Estimates from 20 Years Earlier. J. Infect. Dis. 2021, 224, S558–S567. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Sindhu, K.N.; Kumar, J.S.; Ramasamy, R.K.; Pragasam, A.K.; Aasaithampi, P.; Mohan, V.R.; Kang, G.; John, J. Outbreak of Typhoid Fever in Children of Urban Vellore: A Report from the Surveillance for Enteric Fever in India Cohort. Am. J. Trop. Med. Hyg. 2022, 107, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Tanmoy, A.M.; Andrews, J.R.; Sajib, M.S.I.; Yu, A.T.; Baker, S.; Luby, S.P.; Saha, S.K. Evaluating PCR-Based Detection of Salmonella Typhi and Paratyphi A in the Environment as an Enteric Fever Surveillance Tool. Am. J. Trop. Med. Hyg. 2019, 100, 43–46. [Google Scholar] [CrossRef] [PubMed]
- Achi, C.R.; Ayobami, O.; Mark, G.; Egwuenu, A.; Ogbolu, D.; Kabir, J. Operationalising One Health in Nigeria: Reflections From a High-Level Expert Panel Discussion Commemorating the 2020 World Antibiotics Awareness Week. Front. Public Health 2021, 9, 673504. [Google Scholar] [CrossRef]
- Zhang, Z.; Kermekchiev, M.B.; Barnes, W.M. Direct DNA Amplification from Crude Clinical Samples Using a PCR Enhancer Cocktail and Novel Mutants of Taq. J. Mol. Diagn. 2010, 12, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Aiemjoy, K.; Seidman, J.C.; Saha, S.; Munira, S.J.; Islam Sajib, M.S.; Sium, S.M.A.; Sarkar, A.; Alam, N.; Zahan, F.N.; Kabir, M.S.; et al. Estimating Typhoid Incidence from Community-Based Serosurveys: A Multicohort Study. Lancet Microbe 2022, 3, e578–e587. [Google Scholar] [CrossRef]
- Gunn, J.S.; Marshall, J.M.; Baker, S.; Dongol, S.; Charles, R.C.; Ryan, E.T. Salmonella Chronic Carriage: Epidemiology, Diagnosis, and Gallbladder Persistence. Trends Microbiol. 2014, 22, 648–655. [Google Scholar] [CrossRef] [Green Version]
- Panzner, U.; Mogeni, O.D.; Adu-Sarkodie, Y.; Toy, T.; Jeon, H.J.; Pak, G.D.; Park, S.E.; Enuameh, Y.; Owusu-Dabo, E.; Van Tan, T.; et al. Detection of Salmonella Typhi Nucleic Acid by RT-PCR and Anti-HlyE, -CdtB, -PilL, and -Vi IgM by ELISA at Sites in Ghana, Madagascar and Ethiopia. BMC Infect. Dis. 2022, 22, 766. [Google Scholar] [CrossRef]



| Target | Forward | Reverse |
|---|---|---|
| O-group | Tyv-5′GAG GAA GGG AAA TGA AG C TTT T-3′ Prt-5′CTT GCT ATG GAA GAC ATA ACG AAC C-3′ | 5′-TAG CAA ACT GTC TCC CAC CAT AC-3′ 5′-CGT CTC CAT CAA AAG CTC CAT AGA-3′ |
| H-antigen | H-5′ACT CAG GCT TCC CGT AAC GC-3′ | Hd-5′GGC TAG TAT TGT CCT TAT CGG-3′ Ha-GAG GCC AGC ACC ATC AGT GC |
| N | 120 (%) | |
|---|---|---|
| Sex (%) | Female | 49 (40.8%) |
| Male | 71 (59.2%) | |
| Age (years) | Mean ± S.D | 6.1 ± 2.9 |
| 95% Confidence interval | 5.6–6.6 | |
| Range | 1–14 | |
| Lower quartile | 4 | |
| Median | 6 | |
| Upper quartile | 8 | |
| ≤5 years (%) | 57 (47.5%) | |
| >5 years (%) | 63 (52.5%) |
| Test | Sample Size | The Outcome for S. Typhi |
|---|---|---|
| Salmonella/shigella agar culture | 67 | 23 positives |
| API | 23 | None |
| PCR Target | Number Positive | Number Negative | |
|---|---|---|---|
| Salmonella Typhi O-antigen | tyv | 0 | 47 |
| Salmonella Typhi H-antigen | Hd | 0 | 47 |
| Salmonella Paratyphi O-antigen | prt | 6 | 41 |
| Salmonella Paratyphi H-antigen | Ha | 0 | 41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uwanibe, J.N.; Kayode, T.A.; Oluniyi, P.E.; Akano, K.; Olawoye, I.B.; Ugwu, C.A.; Happi, C.T.; Folarin, O.A. The Prevalence of Undiagnosed Salmonella enterica Serovar Typhi in Healthy School-Aged Children in Osun State, Nigeria. Pathogens 2023, 12, 594. https://doi.org/10.3390/pathogens12040594
Uwanibe JN, Kayode TA, Oluniyi PE, Akano K, Olawoye IB, Ugwu CA, Happi CT, Folarin OA. The Prevalence of Undiagnosed Salmonella enterica Serovar Typhi in Healthy School-Aged Children in Osun State, Nigeria. Pathogens. 2023; 12(4):594. https://doi.org/10.3390/pathogens12040594
Chicago/Turabian StyleUwanibe, Jessica N., Tolulope A. Kayode, Paul E. Oluniyi, Kazeem Akano, Idowu B. Olawoye, Chinedu A. Ugwu, Christian T. Happi, and Onikepe A. Folarin. 2023. "The Prevalence of Undiagnosed Salmonella enterica Serovar Typhi in Healthy School-Aged Children in Osun State, Nigeria" Pathogens 12, no. 4: 594. https://doi.org/10.3390/pathogens12040594





