Natural and Engineered Resistance Mechanisms in Plants against Phytoviruses
Abstract
:1. Introduction
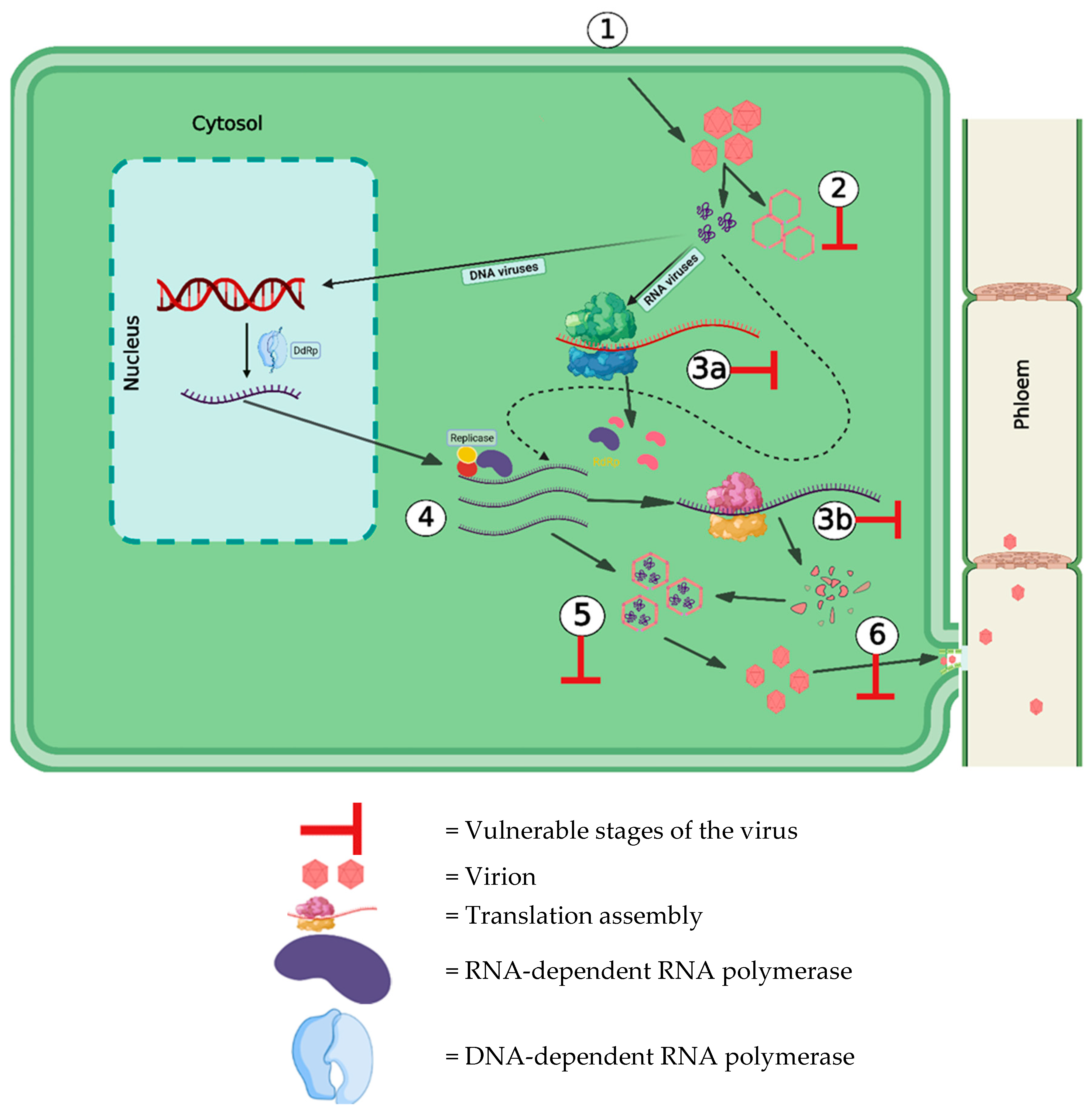
2. Virus Infection Cycle within the Plants
3. Antiviral Defence Mechanisms in Plants
3.1. Natural Resistance to Plant Viruses
3.1.1. Innate Antiviral Immunity
3.1.2. Translation Repression as Virus Resistance
3.1.3. Small RNA-Mediated Antiviral Defence
3.1.4. Dominant Viral Resistance Genes
3.1.5. Resistance to Virus Movement within and between the Cells
3.1.6. Autophagy as Antiviral Mechanism against Plant Viruses
3.1.7. Cross Protection
3.2. Engineered Resistance to Plant Viruses
3.2.1. Pathogen-Derived Resistance
3.2.2. Gene Editing Technologies
Engineering ZFN or TALEN-Based Resistance against Viruses
Engineering CRISPR/CAS-Based Resistance against Plant Viruses
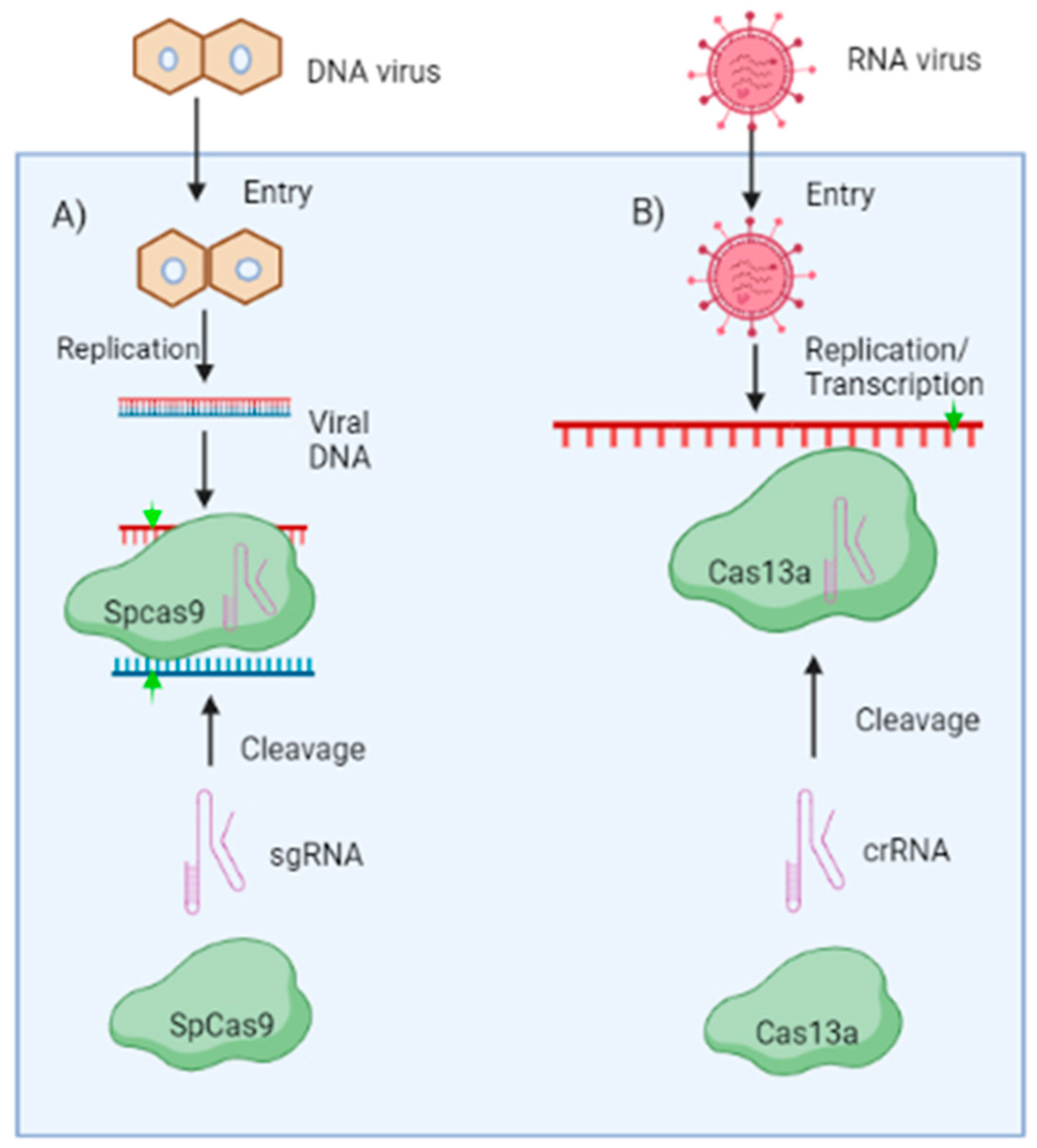
CRISPR/CAS-Based Resistance against DNA Viruses
CRISPR/CAS-Based Resistance against RNA Viruses
| CRISPR/Cas System | Host Plant | Virus | References |
|---|---|---|---|
| SpCas9 | Tobacco and Arabidopsis | Beet-severe curly top virus, Beet curly top virus | [59,61] |
| Tobacco | Bean yellow dwarf virus | [60] | |
| Barley | Wheat dwarf virus | [63,64] | |
| FnCas9 | Tobacco | Tobacco mosaic virus | [65] |
| Tobacco and Arabidopsis | Cucumber mosaic virus | [65] | |
| Tobacco | Tobacco mosaic virus | [66] | |
| LshCas13a | Rice | Southern rice blacked streaked dwarf virus | [66] |
| Tobacco | Turnip mosaic virus | [67] |
4. Reported Resistance Genes (Dominant and/or Recessive) against Plant Viruses in Major Vegetable Crops
5. Conclusions
6. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Soosaar, J.L.; Burch-Smith, T.M.; Dinesh-Kumar, S.P. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005, 3, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Calil, I.P.; Fontes, E.P. Plant immunity against viruses: Antiviral immune receptors in focus. Ann. Bot. 2017, 119, 711–723. [Google Scholar] [CrossRef]
- Schwessinger, B.; Zipfel, C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008, 11, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Scholthof, K.B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738. [Google Scholar] [CrossRef]
- Allan, A.C.; Lapidot, M.; Culver, J.N.; Fluhr, R. An early tobacco mosaic virus-induced oxidative burst in tobacco indicates extracellular perception of the virus coat protein. Plant Physiol. 2001, 126, 97–108. [Google Scholar] [CrossRef]
- Perraki, A.; Gronnier, J.; Gouguet, P.; Boudsocq, M.; Deroubaix, A.F.; Simon, V.; Germain, V. REM1. 3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 2018, 14, e1007378. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D. Plant immune response strategies against pathogens. Plant Arch. 2020, 20, 1169–1174. [Google Scholar]
- Sanfaçon, H. Plant translation factors and virus resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef] [PubMed]
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009, 75, 119–231. [Google Scholar] [PubMed]
- Domashevskiy, A.V.; Cheng, S.Y. Translation Initiation Complex eIFiso4F Targets Pokeweed Antiviral Protein (PAP) to Selectively Depurinate Uncapped Tobacco Etch Virus (TEV) RNA. Biophys. J. 2018, 114, 442. [Google Scholar] [CrossRef]
- Pantaleo, V.; Szittya, G.; Burgyán, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007, 81, 3797–3806. [Google Scholar] [CrossRef]
- Calvo, M.; Martínez-Turiño, S.; García, J.A. Resistance to Plum pox virus strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome-linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant-Microbe Interact. 2014, 27, 1291–1301. [Google Scholar] [CrossRef]
- Ding, S.W.; Han, Q.; Wang, J.; Li, W.X. Antiviral RNA interference in mammals. Curr. Opin. Immunol. 2018, 54, 109–114. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature. 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Llave, C. Virus-derived small interfering RNAs at the core of plant–virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, B.; Sanfaçon, H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 2014, 456, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jia, T.; Chen, X. The ‘how’and ‘where’of plant micro RNAs. New Phytol. 2017, 216, 1002–1017. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Wu, J. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant. 2016, 9, 1302–1314. [Google Scholar] [CrossRef]
- Ranf, S.; Gisch, N.; Schäffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Scheel, D. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433. [Google Scholar] [CrossRef]
- Sugawara, K.; Shiraishi, T.; Yoshida, T.; Fujita, N.; Netsu, O.; Yamaji, Y.; Namba, S. A replicase of potato virus X acts as the resistance-breaking determinant for JAX1-mediated resistance. Mol. Plant-Microbe Interact. 2013, 26, 1106–1112. [Google Scholar] [CrossRef]
- Yoshida, T.; Shiraishi, T.; Hagiwara-Komoda, Y.; Komatsu, K.; Maejima, K.; Okano, Y.; Namba, S. The plant noncanonical antiviral resistance protein JAX1 inhibits potexviral replication by targeting the viral RNA-dependent RNA polymerase. J. Virol. 2019, 93, e01506-18. [Google Scholar] [CrossRef]
- Yoshii, M.; Yoshioka, N.; Ishikawa, M.; Naito, S. Isolation of an Arabidopsis thaliana mutant in which accumulation of cucumber mosaic virus coat protein is delayed. Plant J. 1998, 13, 211–219. [Google Scholar] [CrossRef]
- Gao, Z.; Johansen, E.; Eyers, S.; Thomas, C.L.; Noel Ellis, T.H.; Maule, A.J. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 2004, 40, 376–385. [Google Scholar] [CrossRef]
- Ruffel, S.; Dussault, M.H.; Palloix, A.; Moury, B.; Bendahmane, A.; Robaglia, C.; Caranta, C. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 2002, 32, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Parra, M.A.; Anderberg, R.J.; Carrington, J.C. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 2001, 127, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Fujisaki, K.; Ishikawa, M. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology 2008, 380, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Lukan, T.; Baebler, Š.; Pompe-Novak, M.; Guček, K.; Zagorščak, M.; Coll, A.; Gruden, K. Cell death is not sufficient for the restriction of potato virus Y spread in hypersensitive response-conferred resistance in potato. Front. Plant Sci. 2018, 9, 168. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Codogno, P. The mechanism and physiological function of macroautophagy. J. Innate Immun. 2013, 5, 427–433. [Google Scholar] [CrossRef]
- Espert, L.; Codogno, P.; Biard-Piechaczyk, M. Involvement of autophagy in viral infections: Antiviral function and subversion by viruses. J. Mol. Med. 2007, 85, 811–823. [Google Scholar] [CrossRef]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Liu, Y. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897. [Google Scholar] [CrossRef]
- Li, F.; Zhang, C.; Li, Y.; Wu, G.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1268. [Google Scholar] [CrossRef]
- Hafrén, A.; Üstün, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 2018, 176, 649–662. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Asurmendi, S.; Hopp, H.E.; Beachy, R.N. Tobacco mosaic virus (TMV) and potato virus X (PVX) coat proteins confer heterologous interference to PVX and TMV infection, respectively. J. Gen. Virol. 2006, 87, 1005–1012. [Google Scholar] [CrossRef]
- Yoon, J.Y.; Ahn, H.I.; Kim, M.; Tsuda, S.; Ryu, K.H. Pepper mild mottle virus pathogenicity determinants and cross protection effect of attenuated mutants in pepper. Virus Res. 2006, 118, 23–30. [Google Scholar] [CrossRef]
- Sanford, J.C.; Johnston, S.A. The concept of parasite-derived resistance-deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 1985, 113, 395–405. [Google Scholar] [CrossRef]
- Ferreira, S.A.; Pitz, K.Y.; Manshardt, R.; Zee, F.; Fitch, M.; Gonsalves, D.E. Virus coat protein transgenic papaya provides practical control of papaya ringspot virus in Hawaii. Plant Dis. 2002, 86, 101–105. [Google Scholar] [CrossRef]
- Yang, R.; Xu, H.; Long, M.; Yu, W.; Lu, C.; Wu, G.C.; Chen, Z. Transgenic tomato plants expressing cucumber mosaic virus coat protein and their resistance to CMV. Jiangsu Nong Ye Xue Bao 1995, 11, 40–44. [Google Scholar]
- Zhu, Y.X.; Ou Yang, W.J.; Zhang, Y.F.; Chen, Z.L. Transgenic sweet pepper plants from 1103 Agrobacterium mediated transformation. Plant Cell Rep. 1996, 16, 71–75. [Google Scholar] [CrossRef]
- Kaniewski, W.K.; Thomas, P.E. The potato story. AgBioForum 2004, 7, 41–46. [Google Scholar]
- Davis, M.J.; Ying, Z. Development of papaya breeding lines with transgenic resistance to Papaya ringspot virus. Plant Dis. 2004, 88, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, T.; Cambra, M.; Capote, N.; Zawadzka, B.; Gorris, M.T.; Scorza, R.; Ravelonandro, M. Field trials of plum clones transformed with the Plum pox virus coat protein (PPV-CP) gene. Plant Dis. 2006, 90, 1012–1018. [Google Scholar] [CrossRef]
- Faria, J.C.; Albino, M.M.; Dias, B.B.; Cançado, L.J.; da Cunha, N.B.; Silva, L.D.M.; Aragão, F.J. Partial resistance to Bean golden mosaic virus in a transgenic common bean (Phaseolus vulgaris L.) line expressing a mutated rep gene. Plant Sci. 2006, 171, 565–571. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, Q.; Canzio, D.; Shou, J.; Li, J.; Gorkin, D.U.; Wu, Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 2015, 162, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Ilardi, V.; Tavazza, M. Biotechnological strategies and tools for Plum pox virus resistance: Trans-, intra-, cis-genesis, and beyond. Front. Plant Sci. 2015, 6, 379. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Sera, T. Inhibition of virus DNA replication by artificial zinc finger proteins. J. Virol. 2005, 79, 2614–2619. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Chen, W.; Qian, Y.; Wu, X.; Sun, Y.; Wu, X.; Cheng, X. Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral-conserved nucleotide motif. Virus Genes 2014, 48, 494–501. [Google Scholar] [CrossRef]
- Zaidi, S.S.E.A.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering plant immunity: Using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 2016, 7, 1673. [Google Scholar] [CrossRef]
- Unniyampurath, U.; Pilankatta, R.; Krishnan, M.N. RNA interference in the age of CRISPR: Will CRISPR interfere with RNAi? Int. J. Mol. Sci. 2016, 17, 291. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef] [PubMed]
- Mahas, A.; Mahfouz, M. Engineering virus resistance via CRISPR–Cas systems. Curr. Opin. Virol. 2018, 32, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kis, A.; Hamar, É.; Tholt, G.; Bán, R.; Havelda, Z. Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol. J. 2019, 17, 1004. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Soyars, C.L.; Li, J.; Fei, Q.; He, G.; Peterson, B.A.; Wang, X. CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct. 2018, 2, e00047. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 2018, 16, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.; Ye, J.; Cao, X.; Xu, C.; Chen, B.; Zhou, G. Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol. J. 2019, 17, 1185. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1–9. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Jahn, M.M. Genetics of plant virus resistance. Annu. Rev. Phytopathol. 2005, 43, 581–621. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Maule, A.J.; Caranta, C.; Boulton, M.I. Sources of natural resistance to plant viruses: Status and prospects. Mol. Plant Pathol. 2007, 8, 223–231. [Google Scholar] [CrossRef]
- Tatineni, S.; Hein, G.L. Plant Viruses of Agricultural Importance: Current and Future Perspectives of Virus Disease Management Strategies. Phytopathology 2023, 113, 117–141. [Google Scholar] [CrossRef] [PubMed]
- Zamir, D.; Ekstein-Michelson, I.; Zakay, Y.; Navot, N.; Zeidan, M.; Sarfatti, M.; Czosnek, H. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor. Appl. Genet. 1994, 88, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.; Scott, J.W.; Bai, Y. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA–dependent RNA polymerases. PLoS Genet. 2011, 9, e1003399. [Google Scholar] [CrossRef]
- Ji, Y.; Schuster, D.J.; Scott, J.W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol. Breed. 2007, 20, 271–284. [Google Scholar] [CrossRef]
- Ji, Y.; Scott, J.W.; Schuster, D.J.; Maxwell, D.P. Molecular mapping of Ty-4, a new Tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J. Am. Soc. Hortic. Sci. 2009, 134, 281–288. [Google Scholar] [CrossRef]
- Anbinder, I.; Reuveni, M.; Azari, R.; Paran, I.; Nahon, S.; Shlomo, H.; Levin, I. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009, 119, 519–530. [Google Scholar] [CrossRef]
- Hutton, S.F.; Scott, J.W. Ty-6, a major begomovirus resistance gene located on chromosome 10. TGC Rep. 2014, 64, 14–18. [Google Scholar]
- Spassova, M.I.; Prins, T.W.; Folkertsma, R.T.; Klein-Lankhorst, R.M.; Hille, J.; Goldbach, R.W.; Prins, M. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 2001, 7, 151–161. [Google Scholar] [CrossRef]
- Qi, S.; Shen, Y.; Wang, X.; Zhang, S.; Li, Y.; Islam, M.; Liang, Y. A new NLR gene for resistance to tomato spotted wilt virus in tomato (Solanum lycopersicum). Theor. Appl. Genet. 2022, 135, 1493–1509. [Google Scholar] [CrossRef]
- Lv, J.; Deng, M.; Jiang, S.; Zhu, H.; Li, Z.; Wang, Z.; Zhao, K. Mapping and Function Characterization of the Tomato Spotted wilt Virus Resistance Gene SlCHS3 in Solanum Lycopersicum. Mol. Breed. 2022, 42, 55. [Google Scholar] [CrossRef]
- Pelham, J. Resistance in tomato to tobacco mosaic virus. Euphytica 1966, 15, 258–267. [Google Scholar] [CrossRef]
- Pelham, J. Strain-genotype interaction of tobacco mosaic virus in tomato. Ann. Appl. Biol. 1972, 71, 219–288. [Google Scholar] [CrossRef]
- Sharma, N.; Sahu, P.P.; Prasad, A.; Muthamilarasan, M.; Waseem, M.; Khan, Y.; Prasad, M. The Sw5a gene confers resistance to ToLCNDV and triggers an HR response after direct AC4 effector recognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2101833118. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Kanyuka, K.; Baulcombe, D.C. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 1999, 11, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Bendahmane, A.; Querci, M.; Kanyuka, K.; Baulcombe, D.C. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 2000, 21, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Gebhardt, C.; Bellin, D.; Henselewski, H.; Lehmann, W.; Schwarzfischer, J.; Valkonen, J.P.T. Marker-assisted combination of major genes for pathogen resistance in potato. Theor. Appl. Genet. 2006, 112, 1458–1464. [Google Scholar] [CrossRef]
- Saka, N. A rice (Oryza sativa L.) breeding for field resistance to blast disease (Pyricularia oryzae) in Mountainous Region Agricultural Research Institute, Aichi Agricultural Research Center of Japan. Plant Prod. Sci. 2006, 9, 3–9. [Google Scholar] [CrossRef]
- Fulladolsa, A.C.; Jansky, S.H.; Smith, D.R.; Abramczak, C.M.; Charkowski, A.O. Development and Evaluation of Four Molecular Markers Tightly Linked to the Potato Virus Y Resistance Gene Rychc in Diploid Potato Populations; American Phytopathological Society: St. Paul, MN, USA, 2017. [Google Scholar]
- Kondrák, M.; Kopp, A.; Uri, C.; Sós-Hegedűs, A.; Csákvári, E.; Schiller, M.; Bánfalvi, Z. Mapping and DNA sequence characterisation of the Rysto locus conferring extreme virus resistance to potato cultivar ‘White Lady’. PLoS ONE 2020, 15, e0224534. [Google Scholar] [CrossRef]
- Murphy, J.F.; Blauth, J.R.; Livingstone, K.D.; Lackney, V.K.; Jahn, M.K. Genetic mapping of the pvr1 locus in Capsicum spp. and evidence that distinct potyvirus resistance loci control responses that differ at the whole plant and cellular levels. Mol. Plant-Microbe Interact. 1998, 11, 943–951. [Google Scholar] [CrossRef]
- Yeam, I.; Kang, B.C.; Lindeman, W.; Frantz, J.D.; Faber, N.; Jahn, M.M. Allele-specific CAPS markers based on point mutations in resistance alleles at the pvr1 locus encoding eIF4E in Capsicum. Theor. Appl. Genet. 2005, 112, 178–186. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Frantz, J.D.; Murphy, J.F.; Jahn, M.M. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 2005, 42, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Green, S.K.; Kim, J.S. Sources of Resistance to Viruses of Pepper (Capsicum spp.): A Catalog; (No. 633.8498/G798); AVRDC: Taipei, Taiwan, 1998. [Google Scholar]
- Parrella, G.; Ruffel, S.; Moretti, A.; Morel, C.; Palloix, A.; Caranta, C. Recessive resistance genes against potyviruses are localized in colinear genomic regions of the tomato (Lycopersicon spp.) and pepper (Capsicum spp.) genomes. Theor. Appl. Genet. 2002, 105, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Caranta, C.; Thabuis, A.; Palloix, A. Development of a CAPS marker for the Pvr4 locus: A tool for pyramiding potyvirus resistance genes in pepper. Genome 1999, 42, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Grube, R.C.; Blauth, J.R.; Andrés, A.; Caranta, C.; Jahn, M.K. Identification and comparative mapping of a dominant potyvirus resistance gene cluster in Capsicum. Theor. Appl. Genet. 2000, 101, 852–859. [Google Scholar] [CrossRef]
- Caranta, C.; Palloix, A. Both common and specific genetic factors are involved in polygenic resistance of pepper to several potyviruses. Theor. Appl. Genet. 1996, 92, 15–20. [Google Scholar] [CrossRef]
- Koeda, S.; Onouchi, M.; Mori, N.; Pohan, N.S.; Nagano, A.J.; Kesumawati, E. A recessive gene pepy-1 encoding Pelota confers resistance to begomovirus isolates of PepYLCIV and PepYLCAV in Capsicum annuum. Theor. Appl. Genet. 2021, 134, 2947–2964. [Google Scholar] [CrossRef]
- Pérez-de-Castro, A.; Esteras, C.; Alfaro-Fernández, A.; Daròs, J.A.; Monforte, A.J.; Picó, B.; Gómez-Guillamón, M.L. Fine mapping of wmv1551, a resistance gene to Watermelon mosaic virus in melon. Mol. Breed. 2019, 39, 1–15. [Google Scholar] [CrossRef]
- Gilbert, R.Z.; Kyle, M.M.; Munger, H.M.; Gray, S.M. Inheritance of resistance to Watermelon mosaic virus in Cucumis melo L. HortScience 1994, 29, 107–110. [Google Scholar] [CrossRef]
- Anagnostou, K.; Jahn, M.; Perl-Treves, R. Inheritance and linkage analysis of resistance to zucchini yellow mosaic virus, watermelon mosaic virus, papaya ringspot virus and powdery mildew in melon. Euphytica 2000, 116, 265–270. [Google Scholar] [CrossRef]
- Brotman, Y.; Normantovich, M.; Goldenberg, Z.; Zvirin, Z.; Kovalski, I.; Stovbun, N.; Perl-Treves, R. Dual resistance of melon to Fusarium oxysporum races 0 and 2 and to Papaya ring-spot virus is controlled by a pair of head-to-head-oriented NB-LRR genes of unusual architecture. Mol. Plant 2013, 6, 235–238. [Google Scholar] [CrossRef]
- Pitrat, M.; Lecoq, H. Two alleles for watermelon mosaic virus 1 resistance in melon. Cucurbit Genet. Coop. 1983, 6, 52–53. [Google Scholar]
- Pitrat, M.; Lecoq, H. Inheritance of zucchini yellow mosaic virus resistance in Cucumis melo L. Euphytica 1984, 33, 57–61. [Google Scholar] [CrossRef]
- Danin-Poleg, Y.; Paris, H.S.; Cohen, S.; Rabinowitch, H.D.; Karchi, Z. Oligogenic inheritance of resistance to zucchini yellow mosaic virus in melons. Euphytica 1997, 93, 331–337. [Google Scholar] [CrossRef]
- Dhillon, N.P.S.; Ranjana, R.; Singh, K.; Eduardo, I.; Monforte, A.J.; Pitrat, M.; Singh, P.P. Diversity among landraces of Indian snapmelon (Cucumis melo var. momordica). Genet. Resour. Crop Evol. 2007, 54, 1267–1283. [Google Scholar] [CrossRef]
- Karchi, Z. Inheritance of resistance to cucumber mosaic virus in melons. Phytopathology 1975, 65, 479–481. [Google Scholar] [CrossRef]
- Pascual, L.; Yan, J.; Pujol, M.; Monforte, A.J.; Picó, B.; Martín-Hernández, A.M. CmVPS41 Is a general gatekeeper for resistance to Cucumber mosaic virus phloem entry in melon. Front. Plant Sci. 2019, 10, 1219. [Google Scholar] [CrossRef] [PubMed]
- Essafi, A.; Díaz-Pendón, J.A.; Moriones, E.; Monforte, A.J.; Garcia-Mas, J.; Martín-Hernández, A.M. Dissection of the oligogenic resistance to Cucumber mosaic virus in the melon accession PI 161375. Theor. Appl. Genet. 2009, 118, 275–284. [Google Scholar] [CrossRef]
- Guiu-Aragonés, C.; Monforte, A.J.; Saladié, M.; Corrêa, R.X.; Garcia-Mas, J.; Martín-Hernández, A.M. The complex resistance to cucumber mosaic cucumovirus (CMV) in the melon accession PI161375 is governed by one gene and at least two quantitative trait loci. Mol. Breed. 2014, 34, 351–362. [Google Scholar] [CrossRef]
- Daryono, B.S.; Somowiyarjo, S.; Natsuaki, K.T. New source of resistance to Cucumber mosaic virus in melon. SABRAO J. Breed. Genet. 2003, 35, 19–26. [Google Scholar]
- Pitrat, M.; Wipf-Scheibel, C.; Besombes, D.; Desbiez, C.; Lecoq, H. Resistance of melon to Cucumber Vein Yellowing Virus (CVYV). In Cucurbitaceae 2012, Proceedings of the Xth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Antalya, Turkey, 15–18 October 2012; HAL: Antalya, Turkey, 2012; pp. 157–164. [Google Scholar]
- Romay, G.; Pitrat, M.; Lecoq, H.; Wipf-Scheibel, C.; Millot, P.; Girardot, G.; Desbiez, C. Resistance against melon chlorotic mosaic virus and tomato leaf curl New Delhi virus in melon. Plant Dis. 2019, 103, 2913–2919. [Google Scholar] [CrossRef]
- Sugiyama, M.; Ohara, T.; Sakata, Y. Inheritance of resistance to Cucumber green mottle mosaic virus in Cucumis melo L. ‘Chang Bougi’. J. Jpn. Soc. Hortic. Sci. 2007, 76, 316–318. [Google Scholar] [CrossRef]
- Provvidenti, R. Sources of resistance to viruses in two accessions of Cucumis sativus. Rep. Cucurbit Genet. Coop. USA 1985, 8, 12. [Google Scholar]
- Ramírez-Madera, A.O.; Havey, M.J. Different haplotypes encode the same protein for independent sources of Zucchini Yellow Mosaic Virus resistance in cucumber. HortScience 2017, 52, 1040–1042. [Google Scholar] [CrossRef]
- Cohen, S.; Gertman, E.; Kedar, N. Inheritance of resistance to melon mosaic virus in cucumbers. Phytopathology 1971, 61, 253–255. [Google Scholar] [CrossRef]
- Wai, T.; Grumet, R. Inheritance of resistance to watermelon mosaic virus in the cucumber line TMG-1: Tissue-specific expression and relationship to zucchini yellow mosaic virus resistance. Theor. Appl. Genet. 1995, 91, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Yang, Y.; Zhang, S.; Miao, H.; Lu, H.; Wang, Y.; Gu, X. Genetic analysis and gene mapping of papaya ring spot virus resistance in cucumber. Mol. Breed. 2015, 35, 1–9. [Google Scholar] [CrossRef]
- Grumet, R.; Kabelka, E.; McQueen, S.; Wai, T.; Humphrey, R. Characterization of sources of resistance to the watermelon strain of Papaya ringspot virus in cucumber: Allelism and co-segregation with other potyvirus resistances. Theor. Appl. Genet. 2000, 101, 463–472. [Google Scholar] [CrossRef]
- Kabelka, E.; Grumet, R. Inheritance of resistance to the Moroccan watermelon mosaic virus in the cucumber line TMG-1 and cosegregation with zucchini yellow mosaic virus resistance. Euphytica 1997, 95, 237–242. [Google Scholar] [CrossRef]
- Wal, T.; Staub, J.E.; Kabelka, E.; Grumet, R. Linkage analysis of potyvirus resistance alleles in cucumber. J. Hered. 1997, 88, 454–458. [Google Scholar] [CrossRef]
- Shi, L.; Yang, Y.; Xie, Q.; Miao, H.; Bo, K.; Song, Z.; Gu, X. Inheritance and QTL mapping of cucumber mosaic virus resistance in cucumber (Cucumis sativus L.). PLoS ONE 2018, 13, e0200571. [Google Scholar] [CrossRef]
- Sugiyama, M.; Okuda, M.; Sakata, Y. Evaluation of resistance to melon yellow spot virus in a cucumber germplasm collection. Plant Breed. 2009, 128, 696–700. [Google Scholar] [CrossRef]
- Sugiyama, M.; Kawazu, Y.; Fukino, N.; Yoshioka, Y.; Shimomura, K.; Sakata, Y.; Okuda, M. Mapping of quantitative trait loci for Melon yellow spot virus resistance in cucumber (Cucumis sativus L.). Euphytica 2015, 205, 615–625. [Google Scholar] [CrossRef]
- Eid, S.; Abou-Jawdah, Y.; El-Mohtar, C.; Sobh, H.; Havey, M. Tolerance in Cucumber to Cucurbit yellow stunting disorder virus. Plant Dis. 2006, 90, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Pujol, M.; Alexiou, K.G.; Fontaine, A.S.; Mayor, P.; Miras, M.; Jahrmann, T.; Aranda, M.A. Mapping cucumber vein yellowing virus resistance in cucumber (Cucumis sativus L.) by using BSA-seq analysis. Front. Plant Sci. 2019, 10, 1583. [Google Scholar] [CrossRef]
- Brown, R.N.; Bolanos-Herrera, A.; Myers, J.R.; Miller Jahn, M. Inheritance of resistance to four cucurbit viruses in Cucurbita moschata. Euphytica 2003, 129, 253–258. [Google Scholar] [CrossRef]
- Paris, H.S.; Cohen, S.; Burger, Y.; Yoseph, R. Single-gene resistance to zucchini yellow mosaic virus in Cucurbita moschata. Euphytica 1988, 37, 27–29. [Google Scholar] [CrossRef]
- Paris, H.S.; Cohen, S. Oligogenic inheritance for resistance to zucchini yellow mosaic virus in Cucurbita pepo. Ann. Appl. Biol. 2000, 136, 209–214. [Google Scholar] [CrossRef]
- Wessel-Beaver, L. Cultivar and germplasm release. Release of ‘Soler’tropical pumpkin. J. Agric. Univ. Puerto Rico 2005, 89, 263–266. [Google Scholar]
- Provvidenti, R. New American Summer Squash Cultivars Possessing a High Level of Resistance to a Strata of Zucchini Yellow Mosaic Virus from China. Cucurbit Genet. Coop. Rep. 1997, 20, 57–58. [Google Scholar]
- Gilbert-Albertini, F.; Lecoq, H.; Pitrat, M.; Nicolet, J.L. Resistance of Cucurbita moschata to watermelon mosaic virus type 2 and its genetic relation to resistance to zucchini yellow mosaic virus. Euphytica 1993, 69, 231–237. [Google Scholar] [CrossRef]
- Miranda-Vélez, M.A.; Wessel-Beaver, L.; Rodrigues, J.C.V. Non-transmission of ZYMV and PRSV through resistant Cucurbita moschata genotypes ‘Nigerian Local’and ‘Menina’. Cucurbit Genet. Coop. Rep. 2019, 42, 37. [Google Scholar]
- Saha, D.; Rana, R.S.; Sureja, A.K.; Verma, M.; Arya, L.; Munshi, A.D. Cloning and characterization of NBS-LRR encoding resistance gene candidates from Tomato Leaf Curl New Delhi Virus resistant genotype of Luffa cylindrica Roem. Physiol. Mol. Plant Pathol. 2013, 81, 107–117. [Google Scholar] [CrossRef]
- Shrestha, S.; Michael, V.N.E.; Fu, Y.; Meru, G. Genetic Loci Associated with Resistance to Zucchini Yellow Mosaic Virus in Squash. Plants 2021, 10, 1935. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Pathak, M.; Singla, D.; Sharma, A.; Chhuneja, P.; Sarao, N.K. High-density GBS-based genetic linkage map construction and QTL identification associated with yellow mosaic disease resistance in bitter gourd (Momordica charantia L.). Front. Plant Sci. 2021, 12, 671620. [Google Scholar] [CrossRef] [PubMed]
- Provvidenti, R.; Alconero, R. Sources of resistance to pathotypes of pea seed-borne mosaic virus in the US plant introductions of Pisum sativum. PNL 1988, 20, 30–31. [Google Scholar]
- Smýkal, P.; Šafářová, D.; Navrátil, M.; Dostalová, R. Marker assisted pea breeding: eIF4E allele specific markers to pea seed-borne mosaic virus (PSbMV) resistance. Mol. Breed. 2010, 26, 425–438. [Google Scholar] [CrossRef]
- Ali, M.A. Genetics of resistance to the common bean mosaic virus (bean virus 1) in the bean (Phaseolus vulgaris L.). Phytopathology 1950, 40, 69–79. [Google Scholar]
- Drijfhout, E. Genetic Interaction between Phaseolus vulgaris and Bean Common Mosaic Virus with Implications for Strain Identification and Breeding for Resistance; Wageningen University and Research: Wageningen, The Netherlands, 1978. [Google Scholar]
 : depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.
: depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.
 : depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.
: depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.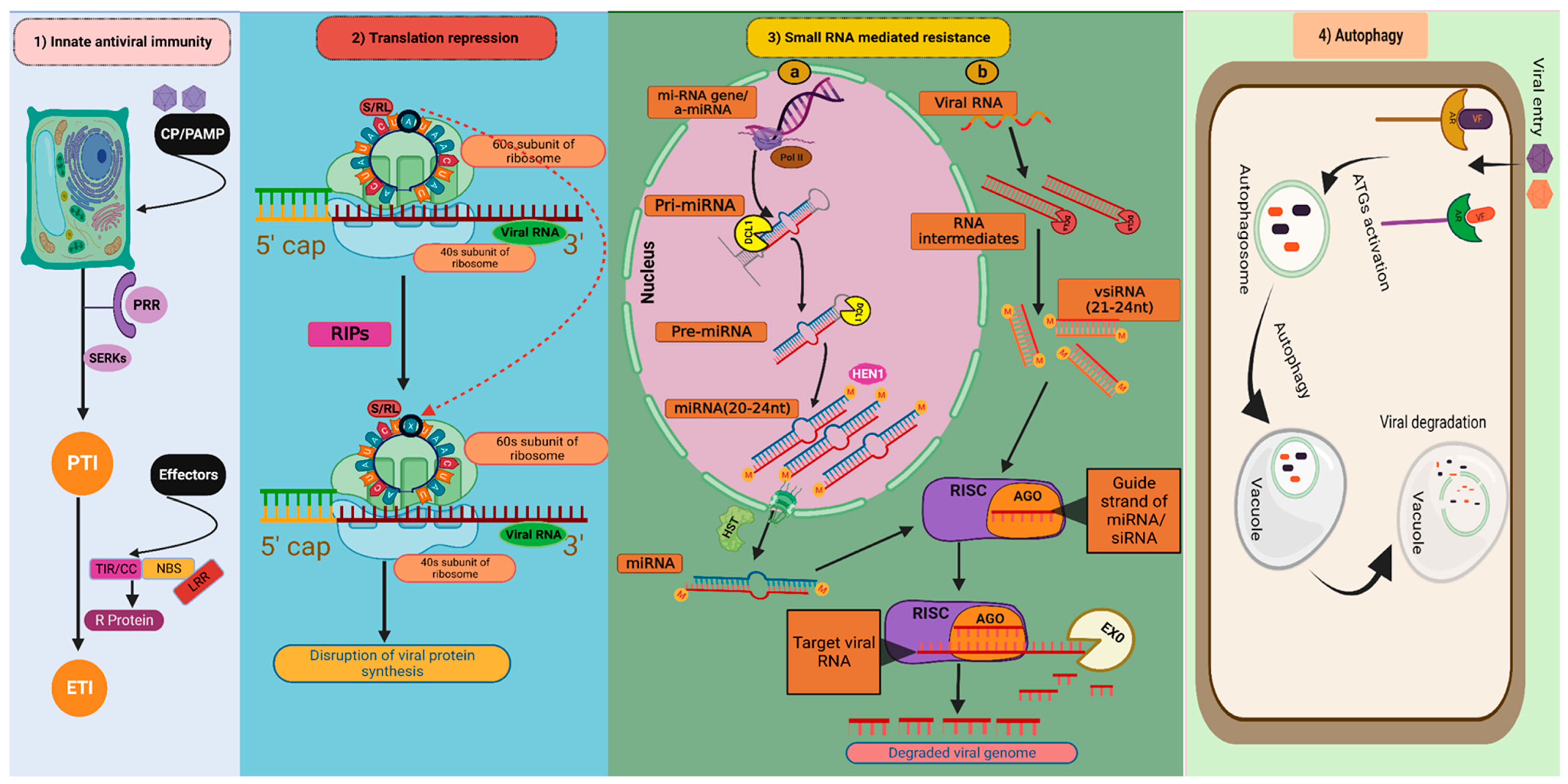
| Crop Species | Target Pathogen | Gene Expressed | Reference |
|---|---|---|---|
| Solanum lycopersicum | Cucumber mosaic virus | Coat protein | [44] |
| Capsicum annum | Cucumber mosaic virus | Coat protein | [45] |
| Solanum tuberosum | Potato leaf roll virus | Coat protein | [46] |
| Carica papaya | Papaya ringspot virus | Coat protein | [47] |
| Prunus domestica | Plum pox virus | Coat protein | [48] |
| Phaseolus vulgaris | Bean golden mosaic virus | +,− RNA of virus replication protein | [49] |
| Carica papaya | Papaya ringspot virus | Viral replicase gene | [50] |
| Prunus domestica | Plum pox virus | Coat protein, P1, HC-Pro | [51] |
| R Gene | Name of the Virus | Reference |
|---|---|---|
| Ty-1 | Tomato yellow leaf curl virus | [72] |
| Ty-2 | Tomato yellow leaf curl virus | [73] |
| Ty-3 | Tomato yellow leaf curl virus | [74] |
| Ty-4 | Tomato yellow leaf curl virus | [75] |
| ty-5 | Tomato yellow leaf curl virus | [76] |
| Ty-6 | Tomato yellow leaf curl virus | [77] |
| Sw-5 | Tomato spotted wilt virus | [78] |
| Sl5R-1 | Tomato spotted wilt virus | [79] |
| SICSH3 | Tomato spotted wilt virus | [80] |
| Tm-1 | Tomato mosaic virus | [81] |
| Tm2 and Tm-22 | Tomato mosaic virus | [82] |
| SlSw5a | Tomato leaf curl New Delhi virus | [83] |
| R Gene | Name of the Virus | Reference |
|---|---|---|
| Rx-1 | Potato virus X | [84] |
| Rx-2 | Potato virus X | [85] |
| Ryadg | Potato virus Y | [86] |
| Y-1 | Potato virus Y | [87] |
| Rychc | Potato virus Y | [88] |
| Ryfsto | Potato virus YNTN | [89] |
| R gene | Name of the Virus | References |
|---|---|---|
| pvr1 | Potato Virus Y, Pepper mottle virus | [90,91] |
| pvr2 | Potato Virus Y | [91,92] |
| pvr3 | Pepper mottle virus | [93,94] |
| Pvr4 | Pepper mottle virus | [95,96] |
| pvr5 | Pepper vein mottle virus | [94] |
| pvr6 | Pepper vein mottle virus | [31,97] |
| Pvr7 | Pepper mottle virus | [96] |
| pepy-1 | Pepper yellow leaf curl Indonesia virus and Pepper yellow leaf curl Aceh virus | [98] |
| Host Plant | Group | R Gene | Name of the Virus | References |
|---|---|---|---|---|
| Cucumis melo | Acidulus | Wmv1551 Wmr | Watermelon mosaic virus | [99,100] |
| Prv2 Prv1 | Papaya ringspot virus | [101,102,103] | ||
| Acidulus | Zym-1 to Zym-3 | Zuchhini yellow mosaic virus | [104,105,106] | |
| Conomon, Conomon | cmv1 cmv1, cmqw3.1, cmqw10.1 Creb-2 | Cucumber mosaic virus | [107,108,109,110,111] | |
| Cvy-1 cvy-2 Cvy-3 | Cucumber vein yellowing virus | [112] | ||
| bgm-1, Bgm-2, Tolcndv | Tomato leaf curl New Delhi virus | [113] | ||
| Makuwa | cgmmv-1, cgmmv-2 | Cucumber green mottle mosaic virus | [114] | |
| Cucumis sativus | zym-1 | Zuchhini yellow mosaic virus | [115,116] | |
| Wmv wmv-2, wmv-3 | Watermelon mosaic virus | [117,118] | ||
| prsv Prsv-2 prsv-1 | Papaya ringspot virus | [119,120,121,122] | ||
| cmv6.1 | Cucumber mosaic virus | [123] | ||
| Swf-1, Swf-2, Swf-3 and Swf-4 | Melon yellow spot virus | [124,125] | ||
| cysdv5.1 | Cucurbit yellow stunting disorder virus | [126] | ||
| Cscys-1 | Cucumber vein yellowing virus | [127] | ||
| Cucurbita moschata | Zym-0, zym-4, Zym-1, Zym-2, Zym-3 zym-6 | Zucchini yellow mosaic virus | [128,129,130,131] | |
| Wmv | Watermelon mosaic virus | [128,132,133] | ||
| prv | Papaya ringspot virus | [128,134] | ||
| cmv | Cucumber mosaic virus | [128,132] | ||
| Luffa cylindrica | RGCLc28 | Tomato leaf curl New Delhi virus | [135] | |
| Cucurbita pepo | QtlZYMV-02 QtlZYMV-04 QtlZYMV-08 QtlZYMV-20 | Zucchini yellow mosaic virus | [136] | |
| Momordica charantia | qYMD.pau_3.1 q.YMD.pau_4.1 qYMD.pau_5.1 | Virus complex | [137] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumdar, A.; Sharma, A.; Belludi, R. Natural and Engineered Resistance Mechanisms in Plants against Phytoviruses. Pathogens 2023, 12, 619. https://doi.org/10.3390/pathogens12040619
Majumdar A, Sharma A, Belludi R. Natural and Engineered Resistance Mechanisms in Plants against Phytoviruses. Pathogens. 2023; 12(4):619. https://doi.org/10.3390/pathogens12040619
Chicago/Turabian StyleMajumdar, Anik, Abhishek Sharma, and Rakesh Belludi. 2023. "Natural and Engineered Resistance Mechanisms in Plants against Phytoviruses" Pathogens 12, no. 4: 619. https://doi.org/10.3390/pathogens12040619
APA StyleMajumdar, A., Sharma, A., & Belludi, R. (2023). Natural and Engineered Resistance Mechanisms in Plants against Phytoviruses. Pathogens, 12(4), 619. https://doi.org/10.3390/pathogens12040619








