The Immune Response in the Uteri and Placentae of Chlamydia abortus-Infected Ewes and Its Association with Pregnancy Outcomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.2. Sample Collection and Storage
2.3. Real-Time PCR Analyses
2.4. Phenotypic Analysis of Inflammatory Cells
2.4.1. Immunohistochemical Labelling of Tissue Sections
2.4.2. Pre-Treatment for Antigen Retrieval IHC
2.4.3. Immunolabelling of Tissue Sections
2.4.4. Determination of the Optimal Concentration of Primary mAb for IHC
| Target | Antigen | Mab Clone | Dilution | Reference |
|---|---|---|---|---|
| Th cells | CD4 | 17D/2 | 1/400 | [35] |
| T cytotoxic | CD8 | SBU-T8 | 1/200 | [35] |
| C. abortus | Chlamydial MOMP | 4/11 | 1/2000 | [33] |
| γδ-T cell | Gamma-delta T cell receptor (TCR) | 86D | 1/100 | [36] |
| γδ-T cell | Gamma-delta workshop cluster-1 (WC1) | CC15 | 1/1000 | [28] |
| NK cells | CD335/NKp46/NCR1 | GR13.1 | 1/200 | [37] |
| TNF-α | TNF-α | CC327 | 1/400 | [32] |
| Treg cells | FoxP3 | FJK-16s | 1/200 | [38] |
| IL-10 | IL-10 | CC318 | 1/100 | [39] |
2.4.5. In Situ Hybridisation
2.4.6. Phenotypical Scoring
2.5. Statistical Analysis
3. Results
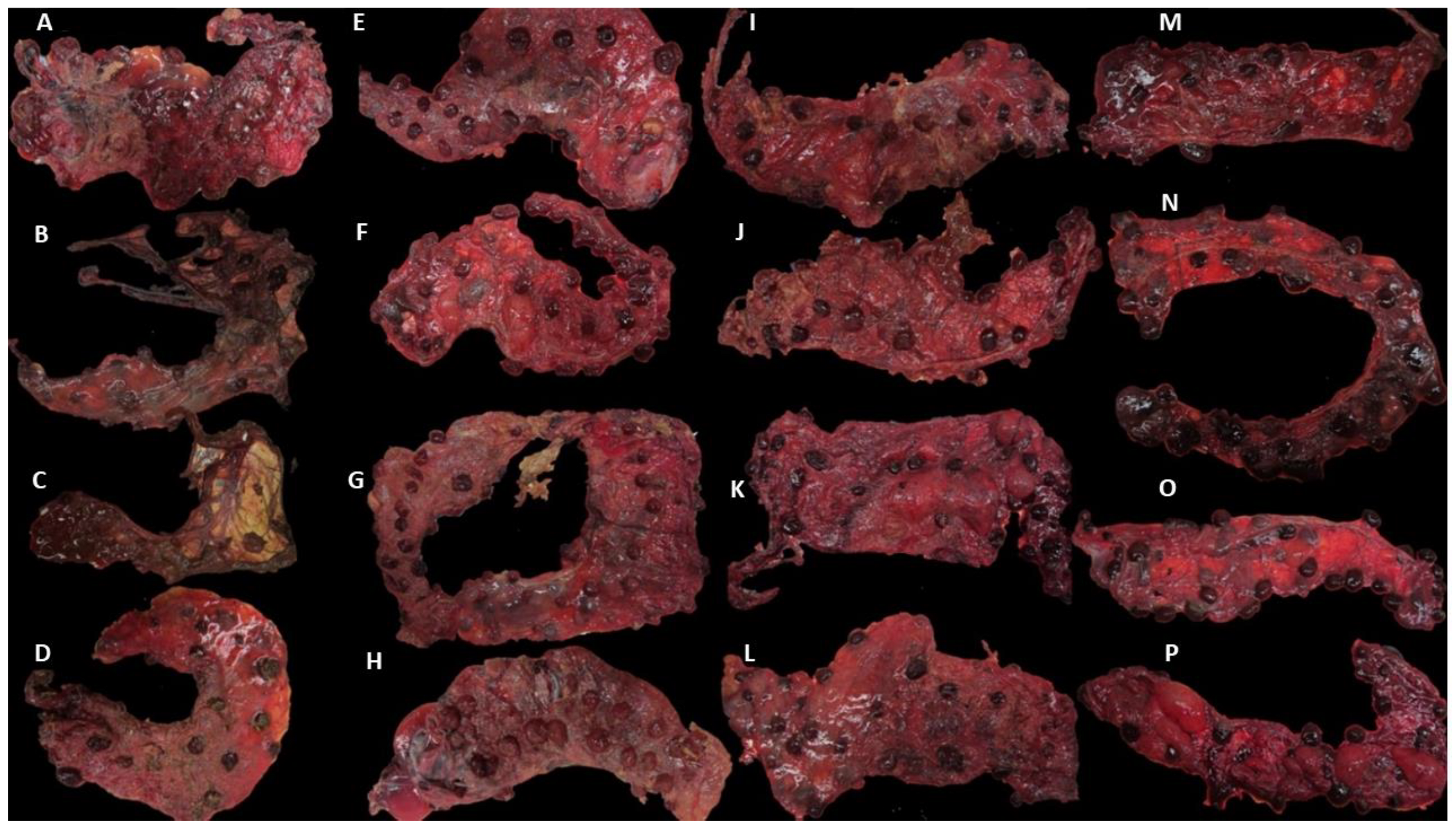
3.1. Clinical Outcomes and Group Allocation
3.2. MZN and qPCR Results in Placentae and Uteri
3.3. Immunolabelling for C. abortus
3.4. Phenotypical Determination of the Cell Infiltration
3.5. In Situ Hybridization for IFN-γ and IL-17A mRNA
3.6. Statistical Analysis of the Immunolabelling and In Situ Hybridisation
3.7. Statistical Analysis of the C. abortus-MOMP Labelling
3.8. Statistical Analysis of the Phenotypical Scoring in Uteri
3.9. Statistical Analysis of the Labelling in Placentae
3.10. Statistical Comparison of the Labelling between the Uteri and the Placentae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Essig, A.; Longbottom, D. Chlamydia abortus: New Aspects of Infectious Abortion in Sheep and Potential Risk for Pregnant Women. Curr. Clin. Microbiol. Rep. 2015, 2, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Longbottom, D.; Coulter, L.J. Animal chlamydioses and zoonotic implications. J. Comp. Pathol. 2003, 128, 217–244. [Google Scholar] [CrossRef]
- Caspe, S.G.; Palarea-Albaladejo, J.; Livingstone, M.; Wattegedera, S.R.; Milne, E.; Sargison, N.D.; Longbottom, D. The extent of placental pathology is negatively correlated to birth weight in ewes infected with the wild-type strain of Chlamydia abortus. Small Rumin. Res. 2023; in press. [Google Scholar] [CrossRef]
- Caspe, S.G.; Livingstone, M.; Frew, D.; Aitchison, K.; Wattegedera, S.R.; Entrican, G.; Palarea-Albaladejo, J.; McNeilly, T.N.; Milne, E.; Sargison, N.D.; et al. The 1B vaccine strain of Chlamydia abortus produces placental pathology indistinguishable from a wild type infection. PLoS ONE 2020, 15, e0242526. [Google Scholar] [CrossRef]
- Livingstone, M.; Wheelhouse, N.; Ensor, H.; Rocchi, M.; Maley, S.; Aitchison, K.; Wattegedera, S.; Wilson, K.; Sait, M.; Siarkou, V.; et al. Pathogenic outcome following experimental infection of sheep with Chlamydia abortus variant strains LLG and POS. PLoS ONE 2017, 12, e0177653. [Google Scholar] [CrossRef] [Green Version]
- Entrican, G.; Buxton, D.; Longbottom, D. Chlamydial infection in sheep: Immune control versus fetal pathology. J. R. Soc. Med. 2001, 94, 273–277. [Google Scholar] [CrossRef] [Green Version]
- Wooding, P.; Burton, G. Placental Immunology, Viviparity, Evolution. In Comparative Placentation: Structures, Functions and Evolution; Wooding, P., Burton, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 231–244. [Google Scholar] [CrossRef]
- Koch, C.A.; Platt, J.L. T cell recognition and immunity in the fetus and mother. Cell Immunol. 2007, 248, 12–17. [Google Scholar] [CrossRef] [Green Version]
- Bainbridge, D.R. Evolution of mammalian pregnancy in the presence of the maternal immune system. Rev. Reprod. 2000, 5, 67–74. [Google Scholar] [CrossRef]
- Wattegedera, S.R.; Doull, L.E.; Goncheva, M.I.; Wheelhouse, N.M.; Watson, D.M.; Pearce, J.; Benavides, J.; Palarea-Albaladejo, J.; McInnes, C.J.; Ballingall, K.; et al. Immunological Homeostasis at the Ovine Placenta May Reflect the Degree of Maternal Fetal Interaction. Front. Immunol. 2019, 9, 3025. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Sung, N.; Gilman-Sachs, A.; Kwak-Kim, J. T Helper (Th) Cell Profiles in Pregnancy and Recurrent Pregnancy Losses: Th1/Th2/Th9/Th17/Th22/Tfh Cells. Front. Immunol. 2020, 11, 2025. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 175, 5–14. [Google Scholar] [CrossRef]
- Wegmann, T.G.; Lin, H.; Guilbert, L.; Mosmann, T.R. Bidirectional cytokine interactions in the maternal-fetal relationship: Is successful pregnancy a TH2 phenomenon? Immunol. Today 1993, 14, 353–356. [Google Scholar] [CrossRef]
- del Rio, L.; Murcia, A.; Buendia, A.J.; Alvarez, D.; Ortega, N.; Navarro, J.A.; Salinas, J.; Caro, M.R. Development of an in vivo model of Chlamydia abortus chronic infection in mice overexpressing IL-10. Vet. Microbiol. 2018, 213, 28–34. [Google Scholar] [CrossRef]
- del Rio, L.; Barberá-Cremades, M.; Navarro, J.A.; Buendía, A.J.; Cuello, F.; Ortega, N.; Gallego, M.C.; Salinas, J.; Caro, M.R. IFN-γ expression in placenta is associated to resistance to Chlamydia abortus after intragastric infection. Microb. Pathog. 2013, 56, 1–7. [Google Scholar] [CrossRef]
- Wattegedera, S.; Sills, K.; Howard, C.J.; Hope, J.C.; McInnes, C.J.; Entrican, G. Variability in cytokine production and cell proliferation by mitogen-activated ovine peripheral blood mononuclear cells: Modulation by interleukin (IL)-10 and IL-12. Vet. Immunol. Immunopathol. 2004, 102, 67–76. [Google Scholar] [CrossRef]
- Seiderer, J.; Elben, I.; Diegelmann, J.; Glas, J.; Stallhofer, J.; Tillack, C.; Pfennig, S.; Jürgens, M.; Schmechel, S.; Konrad, A.; et al. Role of the novel Th17 cytokine IL-17F in inflammatory bowel disease (IBD): Upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm. Bowel Dis. 2007, 14, 437–445. [Google Scholar] [CrossRef]
- Wattegedera, S.; Rocchi, M.; Sales, J.; Howard, C.J.; Hope, J.C.; Entrican, G. Antigen-specific peripheral immune responses are unaltered during normal pregnancy in sheep. J. Reprod. Immunol. 2008, 77, 171–178. [Google Scholar] [CrossRef]
- Entrican, G. Immune regulation during pregnancy and host-pathogen interactions in infectious abortion. J. Comp. Pathol. 2002, 126, 79–94. [Google Scholar] [CrossRef]
- Brown, J.; Howie, S.E.; Entrican, G. A role for tryptophan in immune control of chlamydial abortion in sheep. Vet. Immunol. Immunopathol. 2001, 82, 107–119. [Google Scholar] [CrossRef]
- Leaver, H.A.; Howie, A.; Aitken, I.D.; Appleyard, B.W.; Anderson, I.E.; Jones, G.; Hay, L.A.; Williams, G.E.; Buxton, D. Changes in Progesterone, Oestradiol 17β, and Intrauterine Prostaglandin E2 during Late Gestation in Sheep Experimentally Infected with an Ovine Abortion Strain of Chlamydia psittaci. Microbiol. Soc. 1989, 135, 565–573. [Google Scholar] [CrossRef] [Green Version]
- Fox, A.; Maddox, J.F.; de Veer, M.J.; Meeusen, E.N. γδTCR+ cells of the pregnant ovine uterus express variable T cell receptors and contain granulysin. J. Reprod. Immunol. 2010, 84, 52–56. [Google Scholar] [CrossRef]
- Dogan, S.; Terzioglu, E.; Ucar, S. Innate immune response against HPV: Possible crosstalking with endocervical γδ T cells. J. Reprod. Immunol. 2021, 148, 103435. [Google Scholar] [CrossRef] [PubMed]
- Thomson, N.R.; Yeats, C.; Bell, K.; Holden, M.T.; Bentley, S.D.; Livingstone, M.; Cerdeno-Tarraga, A.M.; Harris, B.; Doggett, J.; Ormond, D.; et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005, 15, 629–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buxton, D.; Anderson, I.E.; Longbottom, D.; Livingstone, M.; Wattegedera, S.; Entrican, G. Ovine chlamydial abortion: Characterization of the inflammatory immune response in placental tissues. J. Comp. Pathol. 2002, 127, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Buxton, D.; Barlow, R.M.; Finlayson, J.; Anderson, I.E.; Mackellar, A. Observations on the pathogenesis of Chlamydia psittaci infection of pregnant sheep. J. Comp. Pathol. 1990, 102, 221–237. [Google Scholar] [CrossRef]
- Wilson, K.; Livingstone, M.; Longbottom, D. Comparative evaluation of eight serological assays for diagnosing Chlamydophila abortus infection in sheep. Vet. Microbiol. 2009, 135, 38–45. [Google Scholar] [CrossRef]
- González, L.; Anderson, I.; Deane, D.; Summers, C.; Buxton, D. Detection of immune system cells in paraffin wax-embedded ovine tissues. J. Comp. Pathol. 2001, 125, 41–47. [Google Scholar] [CrossRef]
- Livingstone, M.; Wheelhouse, N.; Maley, S.W.; Longbottom, D. Molecular detection of Chlamydophila abortus in post-abortion sheep at oestrus and subsequent lambing. Vet. Microbiol. 2009, 135, 134–141. [Google Scholar] [CrossRef]
- Wattegedera, S.R.; Corripio-Miyar, Y.; Pang, Y.; Frew, D.; McNeilly, T.N.; Palarea-Albaladejo, J.; McInnes, C.J.; Hope, J.C.; Glass, E.J.; Entrican, G. Enhancing the toolbox to study IL-17A in cattle and sheep. Vet. Res. 2017, 48, 20. [Google Scholar] [CrossRef] [Green Version]
- Rocchi, M.S.; Bartley, P.M.; Inglis, N.F.; Collantes-Fernandez, E.; Entrican, G.; Katzer, F.; Innes, E.A. Selection of Neospora caninum antigens stimulating bovine CD4+ve T cell responses through immuno-potency screening and proteomic approaches. Vet. Res. 2011, 42, 91. [Google Scholar] [CrossRef] [Green Version]
- Wheelhouse, N.; Wattegedera, S.; Stanton, J.; Maley, S.; Watson, D.; Jepson, C.; Deane, D.; Buxton, D.; Longbottom, D.; Baszler, T.; et al. Ovine trophoblast is a primary source of TNFalpha during Chlamydophila abortus infection. J. Reprod. Immunol. 2009, 80, 49–56. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, M.C.; Herring, A.J.; Andersen, A.A.; Jones, G.E. Electrophoretic analysis of the major outer membrane protein of Chlamydia psittaci reveals multimers which are recognized by protective monoclonal antibodies. Infect. Immun. 1995, 63, 2387–2389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannitti, F.; Anderson, M.; Miller, M.; Rowe, J.; Sverlow, K.; Vasquez, M.; Cantón, G. Chlamydia pecorum: Fetal and placental lesions in sporadic caprine abortion. J. Vet. Diagn. Investig. 2016, 28, 184–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddox, J.F.; Mackay, C.R.; Brandon, M.R. Surface antigens, SBU-T4 and SBU-T8, of sheep T lymphocyte subsets defined by monoclonal antibodies. Immunology 1985, 55, 739–748. [Google Scholar]
- Davis, W.C.; Brown, W.C.; Hamilton, M.J.; Wyatt, C.R.; Orden, J.A.; Khalid, A.M.; Naessens, J. Analysis of monoclonal antibodies specific for the gamma delta TcR. Vet. Immunol. Immunopathol. 1996, 52, 275–283. [Google Scholar] [CrossRef]
- Connelley, T.; Storset, A.K.; Pemberton, A.; MacHugh, N.; Brown, J.; Lund, H.; Morrison, I.W. NKp46 defines ovine cells that have characteristics corresponding to NK cells. Vet. Res. 2011, 42, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeilly, T.N.; McIntyre, J.; Frew, D.; Griffiths, D.J.; Wattegedera, S.R.; van den Broek, A.; Huntley, J.F. Infestation of sheep with Psoroptes ovis, the sheep scab mite, results in recruitment of Foxp3+ T cells into the dermis. Parasite Immunol. 2010, 32, 361–369. [Google Scholar] [CrossRef]
- Kwong, L.S.; Hope, J.C.; Thom, M.L.; Sopp, P.; Duggan, S.; Bembridge, G.P.; Howard, C.J. Development of an ELISA for bovine IL-10. Vet. Immunol. Immunopathol. 2002, 85, 213–223. [Google Scholar] [CrossRef]
- Anderson, I.E.; Reid, H.W.; Nettleton, P.F.; McInnes, C.J.; Haig, D.M. Detection of cellular cytokine mRNA expression during orf virus infection in sheep: Differential interferon-gamma mRNA expression by cells in primary versus reinfection skin lesions. Vet. Immunol. Immunopathol. 2001, 83, 161–176. [Google Scholar] [CrossRef]
- Caspe, S.G.; Palarea-Albaladejo, J.; Underwood, C.; Livingstone, M.; Wattegedera, S.R.; Milne, E.; Sargison, N.D.; Chianini, F.; Longbottom, D. Distribution and Severity of Placental Lesions Caused by the Chlamydia abortus 1B Vaccine Strain in Vaccinated Ewes. Pathogens 2021, 10, 543. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Brooks, M.E.; Kristensen, K.; van Benthem, K.J.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Machler, M.; Bolker, B.M. glmmTMB Balances Speed and Flexibility Among Packages for Zero-inflated Generalized Linear Mixed Modeling. R J. 2017, 9, 378–400. [Google Scholar] [CrossRef] [Green Version]
- Lenth, R.V. Emmeans: Estimated Marginal Means, aka Least-Squares Means. R Package Version 1.7.3. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 7 May 2023).
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks, CA, USA, 2019. [Google Scholar]
- Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R Package Version 0.4.5. Available online: https://CRAN.R-project.org/package=DHARMa (accessed on 7 May 2023).
- Lüdecke, D.; Ben-Shachar, M.; Patil, I.; Waggoner, P.; Makowski, D. performance: An R Package for Assessment, Comparison and Testing of Statistical Models. J. Open Source Softw. 2021, 6, 3139. [Google Scholar] [CrossRef]
- Entrican, G.; Wattegedera, S.R.; Griffiths, D.J. Exploiting ovine immunology to improve the relevance of biomedical models. Mol. Immunol. 2015, 66, 68–77. [Google Scholar] [CrossRef]
- Wooding, F.B.P.; Flint, A.P.F. Placentation; Springer: Dordrecht, The Netherlands, 1994; pp. 233–460. [Google Scholar] [CrossRef]
- Longbottom, D.; Psarrou, E.; Livingstone, M.; Vretou, E. Diagnosis of ovine enzootic abortion using an indirect ELISA (rOMP91B iELISA) based on a recombinant protein fragment of the polymorphic outer membrane protein POMP91B of Chlamydophila abortus. FEMS Microbiol. Lett. 2001, 195, 157–161. [Google Scholar] [CrossRef]
- Rank, R.G.; Bowlin, A.K.; Kelly, K.A. Characterization of lymphocyte response in the female genital tract during ascending Chlamydial genital infection in the guinea pig model. Infect. Immun. 2000, 68, 5293–5298. [Google Scholar] [CrossRef] [Green Version]
- Montes de Oca, R.; Buendia, A.J.; Del Rio, L.; Sanchez, J.; Salinas, J.; Navarro, J.A. Polymorphonuclear Neutrophils Are Necessary for the Recruitment of CD8+ T Cells in the Liver in a Pregnant Mouse Model of Chlamydophila abortus (Chlamydia psittaci Serotype 1) Infection. Infect. Immun. 2000, 68, 1746–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- del Rio, L.; Buendia, A.J.; Sánchez, J.; Garcés, B.; Caro, M.R.; Gallego, M.C.; Bernabé, A.; Cuello, F.; Salinas, J. Chlamydophila abortus (Chlamydia psittaci serotype 1) Clearance is Associated with the Early Recruitment of Neutrophils and CD8+T Cells in a Mouse Model. J. Comp. Pathol. 2000, 123, 171–181. [Google Scholar] [CrossRef]
- Navarro, J.A.; García de la Fuente, J.N.; Sánchez, J.; Martínez, C.M.; Buendía, A.J.; Gutiérrez-Martín, C.B.; Rodriguez-Ferri, E.F.; Ortega, N.; Salinas, J. Kinetics of Infection and Effects on the Placenta of Clamydophila abortus in Experimentally Infected Pregnant Ewes. Vet. Pathol. 2004, 41, 498–505. [Google Scholar] [CrossRef]
- Kronenberg, M.; Havran, W.L. Frontline T cells: Gamma delta T cells and intraepithelial lymphocytes. Immunol. Rev. 2007, 215, 5–7. [Google Scholar] [CrossRef]
- Prinz, I.; Silva-Santos, B.; Pennington, D.J. Functional development of γδ T cells. Eur. J. Immunol. 2013, 43, 1988–1994. [Google Scholar] [CrossRef] [PubMed]
- Bartoskova, A.; Turanek-Knotigova, P.; Matiasovic, J.; Oreskovic, Z.; Vicenova, M.; Stepanova, H.; Ondrackova, P.; Vitasek, R.; Leva, L.; Moore, P.F. γδ T lymphocytes are recruited into the inflamed uterus of bitches suffering from pyometra. Vet. J. 2012, 194, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Fiorenza, M.F.; Amaral, C.D.S.; da Anunciação, A.R.A.; Portela, V.V.M.; Marey, M.A.; Miyamoto, A.; Antoniazzi, A.Q. Possible impact of neutrophils on immune responses during early pregnancy in ruminants. Anim. Reprod. 2021, 18, e20210048. [Google Scholar] [CrossRef] [PubMed]
- Stewart, I.; Peel, S. Mouse Granulated Metrial Gland Cell Cytotoxicity of Wehi 164 Cells: Is There a Role for Interleukin-3 and Tumour Necrosis Factor-α? Am. J. Reprod. Immunol. 1999, 41, 423–427. [Google Scholar] [CrossRef]
- Kinsky, R.; Delage, G.; Rosin, N.; Thang, M.N.; Hoffmann, M.; Chaouat, G. A murine model of NK cell mediated resorption. Am. J. Reprod. Immunol. 1990, 23, 73–77. [Google Scholar] [CrossRef]
- Tekin, Ş.; Hansen, P.J. Natural Killer-Like Cells in the Sheep: Functional Characterization and Regulation by Pregnancy-Associated Proteins. Exp. Biol. Med. 2002, 227, 803–811. [Google Scholar] [CrossRef]
- Tuo, W.; Ott, T.L.; Bazer, F.W. Natural killer cell activity of lymphocytes exposed to ovine, type I, trophoblast interferon. Am. J. Reprod. Immunol. 1993, 29, 26–34. [Google Scholar] [CrossRef]
- Buendía, A.J.; Martínez, C.M.; Ortega, N.; Del Río, L.; Caro, M.R.; Gallego, M.C.; Sánchez, J.; Navarro, J.A.; Cuello, F.; Salinas, J. Natural killer (NK) cells play a critical role in the early innate immune response to Chlamydophila abortus infection in mice. J. Comp. Pathol. 2004, 130, 48–57. [Google Scholar] [CrossRef]
- Entrican, G.; Wattegedera, S.; Rocchi, M.; Wheelhouse, N. Pregnancy, indoleamine 2,3-dioxygenase (IDO) and chlamydial abortion: An unresolved paradox. Vet. Microbiol. 2009, 135, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Dubin, P.J.; Kolls, J.K. Th17 cytokines and mucosal immunity: New Paradigms in Inflammation. Immunol. Rev. 2008, 226, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Helfrich, A.L.; Reichenbach, H.-D.; Meyerholz, M.M.; Schoon, H.-A.; Arnold, G.J.; Fröhlich, T.; Weber, F.; Zerbe, H. Novel sampling procedure to characterize bovine subclinical endometritis by uterine secretions and tissue. Theriogenology 2020, 141, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Immaculate Mbongo, L.; Yamunah Devi, A.; Zain, S.; Omar, S.Z.; Mohamed, Z. Protein Profiling of Women with Spontaneous Preterm Birth. Pharmacology 2015, 96, 44–48. [Google Scholar] [CrossRef] [PubMed]






| Probe | Reactivity | NCBI Accession No | Reference |
|---|---|---|---|
| RNAscopeTM Probe-Oa-IL17A-C1 | IL-17A | XM_004018887.4 | [30] |
| RNAscopeTM Probe-Oa-IFNG-XCh | IFN-γ | NM_001009803.1 | [40] |
| Animal ID | No | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | DD | DD | DD | DL | DL | DL | LL | LL | NC | NC | |
| Macroscopic lesions (%) | H1 | 90 | 100 | 100 | 70 | 30 | 100 | 20 | 10 | 0 | 0 |
| H2 | 25 | 20 | 100 | 30 | 90 | 100 | 10 | 40 | 0 | 0 | |
| mZN | H1C1 | +++ | ++ | + | ++ | + | ++ | +++ | ++ | − | − |
| H1C2 | ++ | ++ | +++ | ++ | + | ++ | +++ | ++ | − | − | |
| H2C1 | ++ | + | ++ | + | ++ | +++ | +++ | ++ | − | − | |
| H2C2 | ++ | + | ++ | + | ++ | +++ | +++ | ++ | − | − | |
| qPCR | H1C1 | 2.10 × 104 | 1.70 × 104 | 1.30 × 103 | 1.70 × 104 | 2.10 × 103 | 1.70 × 104 | 3.20 × 105 | 1.20 × 104 | 0 | 2.7 |
| H1C2 | 3.00 × 103 | 1.10 × 104 | 1.00 × 104 | 5.90 × 103 | 2.70 × 103 | 2.80 × 104 | 1.60 × 102 | 3.50 × 104 | 2 | 1 | |
| H2C2 | 8.30 × 103 | 5.90 × 102 | 9.20 × 103 | 1.90 × 103 | 5.60 × 104 | 1.70 × 105 | 5.10 × 104 | 1.00 × 104 | 1.5 | 8.8 | |
| H2C2 | 5.50 × 104 | 8.8 | 4.20 × 104 | 1.10 × 103 | 5.70 × 104 | 1.00 × 105 | 1.30 × 105 | 3.70 × 103 | 8.5 | 3.2 | |
| Macroscopic lesions (%) | P1 | 60 | 20 | 80 | 40 | 90 | NF | 80 | 10 | 0 | 0 |
| P2 | NF | 40 | NF | 0 | 25 | 100 | 20 | 40 | 0 | 0 | |
| mZN | P1 Px | ++ | ++ | ++ | ++ | +++ | NF | +++ | +++ | − | − |
| P1d | ++ | + | + | ++ | +++ | NF | ++ | +++ | − | − | |
| P2 Px | NF | +++ | NF | + | ++ | ++ | +++ | +++ | − | − | |
| P2D | NF | +++ | NF | + | +++ | ++ | +++ | +++ | − | − | |
| qPCR | P1Px | 2.10 × 105 | 9.00 × 103 | 4.30 × 106 | 3.00 × 105 | 2.00 × 106 | NF | 5.50 × 106 | 1.40 × 107 | 170 | 45 |
| P1D | 6.40 × 105 | 3.90 × 103 | 2.30 × 106 | 1.00 × 105 | 2.00 × 106 | NF | 5.60 × 106 | 1.10 × 107 | 120 | 41 | |
| P2Px | NA | 2.30 × 106 | NA | 1.30 × 103 | 2.40 × 105 | 1.00 × 107 | 1.20 × 107 | 2.40 × 106 | 370 | 48 | |
| P2D | NA | 8.80 × 105 | NA | 9.10 × 104 | 2.70 × 104 | 3.20 × 106 | 8.50 × 106 | 1.10 × 107 | 170 | 41 | |
| Swabs | 1.80 × 106 | 3.50 × 106 | 4.30 × 106 | 6.50 × 105 | 2.50 × 106 | 8.70 × 106 | 1.80 × 105 | 2.90 × 106 | 40 | 20 |
| Tissue | Uteri | Placentae | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | DD | DL | LL | NC | DD | DL | LL | NC | |
| MOMP | Q1 | 0.75 | 0 | 0 | 0 | 5 | 2 | 12.75 | 0 |
| MED | 5 | 10 | 1 | 0 | 24 | 11.5 | 24.5 | 0 | |
| Q3 | 16 | 22 | 8.3 | 0 | 46.25 | 27.5 | 38.75 | 0 | |
| CD4 | Q1 | 3.75 | 7.25 | 3 | 1 | 0 | 0 | 0 | 0 |
| MED | 11 | 19 | 6 | 3 | 0 | 0 | 0 | 0 | |
| Q3 | 70.25 | 47.75 | 14.25 | 6.5 | 0 | 0 | 0 | 0 | |
| CD8 | Q1 | 3.75 | 9.25 | 3 | 1 | 0 | 0 | 0 | 0 |
| MED | 11.5 | 22.5 | 8.5 | 4 | 0 | 0 | 0 | 0 | |
| Q3 | 35 | 71.5 | 19 | 6.25 | 0 | 0 | 0 | 0 | |
| γδ-TCR | Q1 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 0 |
| MED | 3 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | |
| Q3 | 6 | 6 | 6 | 2.25 | 0 | 0 | 0 | 0 | |
| γδ-WC1 | Q1 | 7.75 | 12 | 10 | 3.75 | 0 | 0 | 0 | 0 |
| MED | 16 | 20 | 17 | 7.5 | 0 | 0 | 0 | 0 | |
| Q3 | 29.5 | 45.75 | 26 | 15 | 0 | 1 | 0 | 0 | |
| NKp46 | Q1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| MED | 2 | 3 | 2 | 1 | 0 | 0 | 0 | 0 | |
| Q3 | 6 | 7 | 4 | 2 | 0 | 0 | 0 | 0 | |
| TNF-α | Q1 | 2 | 11.3 | 2 | 0 | 8 | 0 | 0 | 0 |
| MED | 24 | 28 | 8.5 | 1 | 20.5 | 3 | 2 | 0 | |
| Q3 | 85 | 41 | 50 | 4 | 51 | 15.25 | 15 | 5 | |
| IL-10 | Q1 | 3 | 6 | 2.75 | 4 | 0 | 0 | 0 | 0 |
| MED | 6.5 | 9 | 5 | 6.5 | 0 | 2 | 1 | 0 | |
| Q3 | 14.25 | 15 | 7.25 | 9 | 1 | 4 | 3 | 1 | |
| FoxP3 | Q1 | 4.75 | 3 | 7 | 3.75 | 0 | 6 | 2 | 2 |
| MED | 11 | 6.5 | 10.5 | 8 | 0 | 9 | 5 | 4 | |
| Q3 | 18.25 | 12 | 14 | 14.25 | 1 | 13.75 | 8 | 7 | |
| IFN-γ | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MED | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 1 | 3 | 1 | 0 | 0 | 1 | 0 | 0 | |
| IL-17A | Q1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MED | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Q3 | 1 | 0 | 0 | 0 | 0.25 | 0 | 0 | 0 | |
| Tissue | Uteri | Placentae | ||||||
|---|---|---|---|---|---|---|---|---|
| Group | DD | DL | LL | NC | DD | DL | LL | NC |
| MOMP | 6.4 a (2.7–15.6) | 14.2 a (6.0–34.0) | 4.1 a (1.4–12.1) | - | 27.1 a (18.3–40.1) | 18.0 a (12.7–25.5) | 27.5 a (18.6–40.6) | - |
| CD4 T cells | 24.3 a (10.0–58.9) | 32.7 a (13.5–80.0) | 11.8 a (4.0–34.8) | 5.4 a (1.8–15.9) | 3.1 a (1.8–5.3) | 0.3 b (0.1–0.9) | 0.9 b (0.4–2.0) | 0.4 b (0.1–1.2) |
| CD8 T cells | 15.2 ab (6.5–35.5) | 40.6 b (17.4–94.8) | 9.8 ab (3.5–27.8) | 4.1 a (1.5–11.7) | - | - | - | - |
| γδ- TCR | 2.9 a (1.5–5.7) | 5.0 a (2.6–9.8) | 4.4 a (1.9–9.9) | 1.2 a (0.5–2.8) | - | - | - | - |
| γδ-WC1 | 20.3 ab (12.1–34.2) | 34.0 b (20.2–57.2) | 18.7 ab (9.9–35.3) | 8.8 a (4.6–16.6) | - | - | - | - |
| NKp46 | 3.3 ab (1.7–6.4) | 6.7 b (3.5–12.8) | 2.8 ab (1.2–6.1) | 1.2 a (0.5–2.7) | - | - | - | - |
| TNF-α | 23.2 b (7.3–73.2) | 34.4 b (10.9–108.7) | 20.0 ab (4.9–81.6) | 2.5 a (0.6–10.0) | 35.4 a (19.0–66.1) | 10.9 b (6.4–18.5) | 12.6 b (7.0–22.6) | 6.6 b (3.6–12.2) |
| IL-10 | 8.7 a (4.6–16.1) | 12.6 a (6.8–23.5) | 5.1 a (2.4–10.9) | 6.4 a (3.0–13.8) | 2.8 a (2.1–4.0) | 3.7 a (3.1–4.3) | 3.2 a (2.5–4.1) | 0.7 b (0.5–1.0) |
| FoxP3 | 8.5 a (4.9–15.0) | 9.0 a (5.1–15.4) | 10.5 a (5.4–20.4) | 8.4 a (4.3–16.4) | 2.3 a (1.3–4.1) | 10.2 b (7.1–14.5) | 5.6 bc (3.5–9.0) | 4.3 ac (2.8–6.6) |
| IFN-γ | 1.7 a (0.6–5.1) | 2.2 a (0.7–6.3) | 1.0 a (0.3–1.5) | - | 0.4 ab (0.2–0.8) | 0.6 b (0.3–1.0) | 0.3 ab (0.2–0.6) | 0.1 a (0.1–0.3) |
| IL-17A | 0.5 a (0.3–0.9) | 0.2 a (0.1–0.4) | 0.2 a (0.1–0.4) | 0.2 a (0.1–0.4) | 0.3 a (0.1–0.6) | 0.1 a (0.0–0.2) | 0.0 a (0.0–0.1) | 0.3 a (0.1–0.6) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caspe, S.G.; Ewing, D.A.; Livingstone, M.; Underwood, C.; Milne, E.; Sargison, N.D.; Wattegedera, S.R.; Longbottom, D. The Immune Response in the Uteri and Placentae of Chlamydia abortus-Infected Ewes and Its Association with Pregnancy Outcomes. Pathogens 2023, 12, 846. https://doi.org/10.3390/pathogens12060846
Caspe SG, Ewing DA, Livingstone M, Underwood C, Milne E, Sargison ND, Wattegedera SR, Longbottom D. The Immune Response in the Uteri and Placentae of Chlamydia abortus-Infected Ewes and Its Association with Pregnancy Outcomes. Pathogens. 2023; 12(6):846. https://doi.org/10.3390/pathogens12060846
Chicago/Turabian StyleCaspe, Sergio Gaston, David Andrew Ewing, Morag Livingstone, Clare Underwood, Elspeth Milne, Neil Donald Sargison, Sean Ranjan Wattegedera, and David Longbottom. 2023. "The Immune Response in the Uteri and Placentae of Chlamydia abortus-Infected Ewes and Its Association with Pregnancy Outcomes" Pathogens 12, no. 6: 846. https://doi.org/10.3390/pathogens12060846
APA StyleCaspe, S. G., Ewing, D. A., Livingstone, M., Underwood, C., Milne, E., Sargison, N. D., Wattegedera, S. R., & Longbottom, D. (2023). The Immune Response in the Uteri and Placentae of Chlamydia abortus-Infected Ewes and Its Association with Pregnancy Outcomes. Pathogens, 12(6), 846. https://doi.org/10.3390/pathogens12060846






