Functional Granulocyte–Macrophage Colony-Stimulating Factor (GM-CSF) Delivered by Canine Histiocytic Sarcoma Cells Persistently Infected with Engineered Attenuated Canine Distemper Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Virus Neutralization
2.3. Virus Titration
2.4. Immunofluorescence
2.5. RNA Isolation and cDNA Synthesis
2.6. Primer Design
2.7. Reverse Transcription Quantitative PCR (RT-qPCR)
2.8. Immunoblotting
2.9. Immunohistochemistry
2.10. Scratch Wound Assay
2.11. Cell Duplication Assay
(number of counted cells × 0.33)/3 × 0.1 mm3 = number of cells/mL
2.12. Statistical Analysis
3. Results
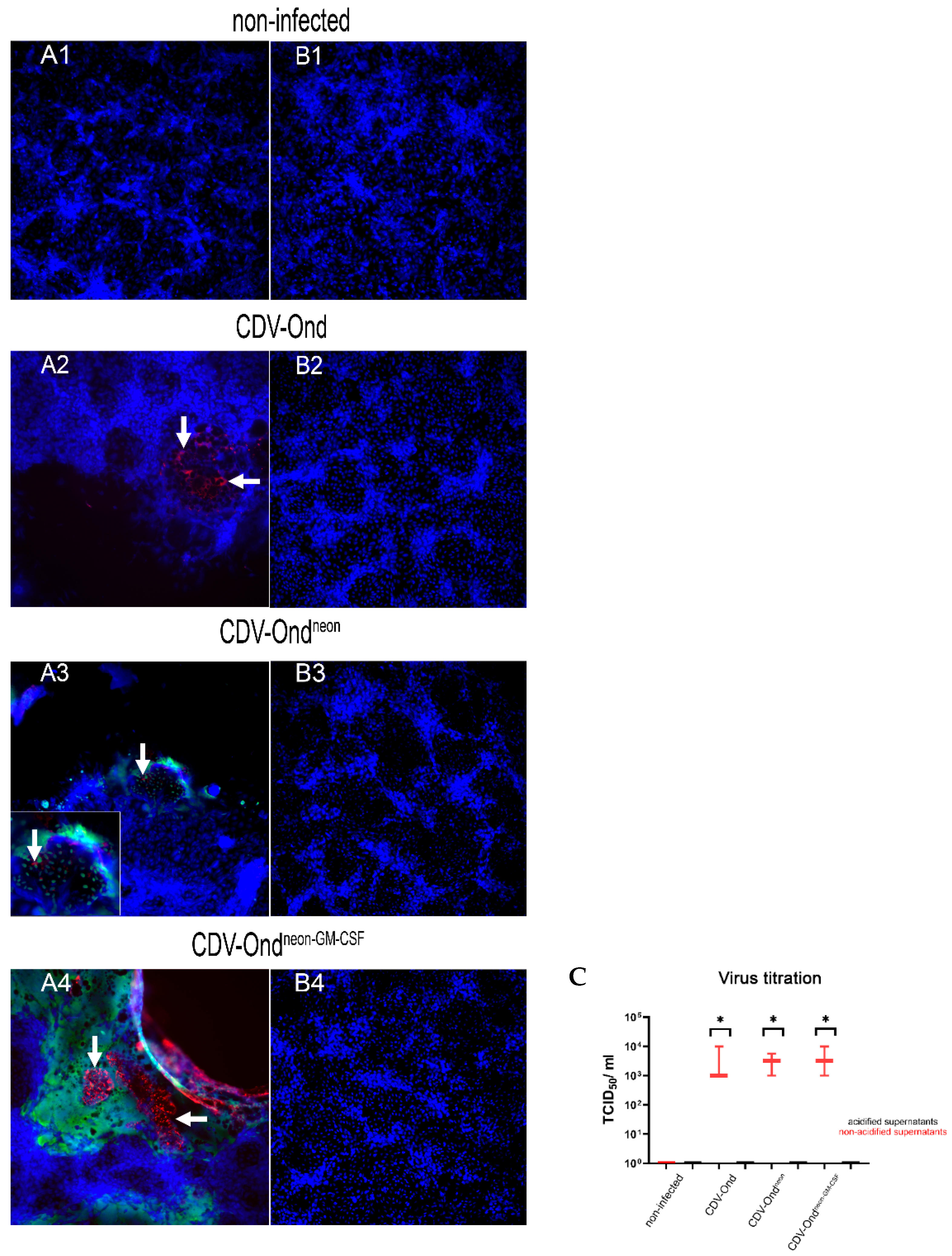
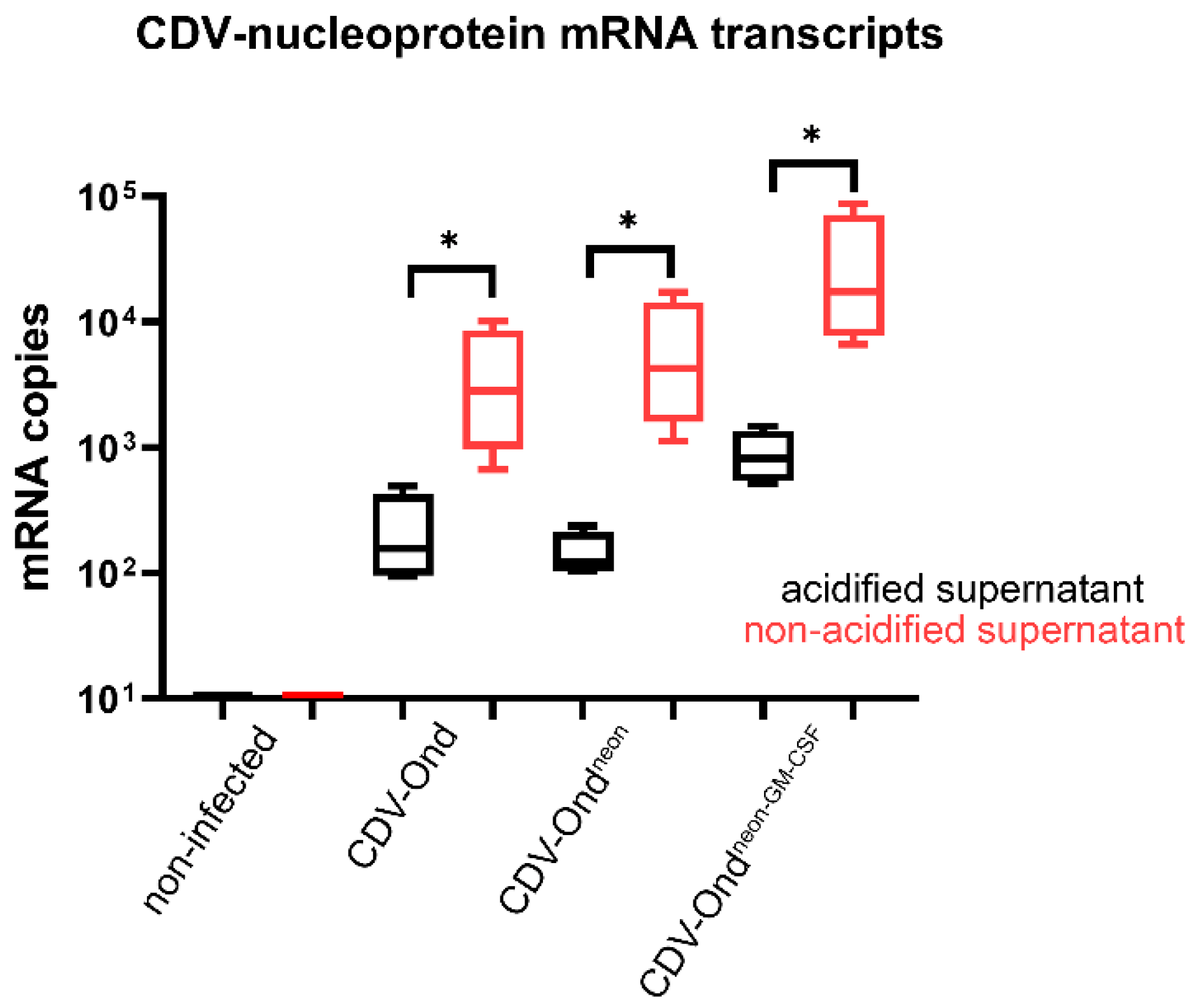
3.1. Acidic Inactivation of the Supernatant Results in CDV Neutralization
3.2. Acidic Inactivation of the Supernatant Does Not Affect the GM-CSF Protein Content
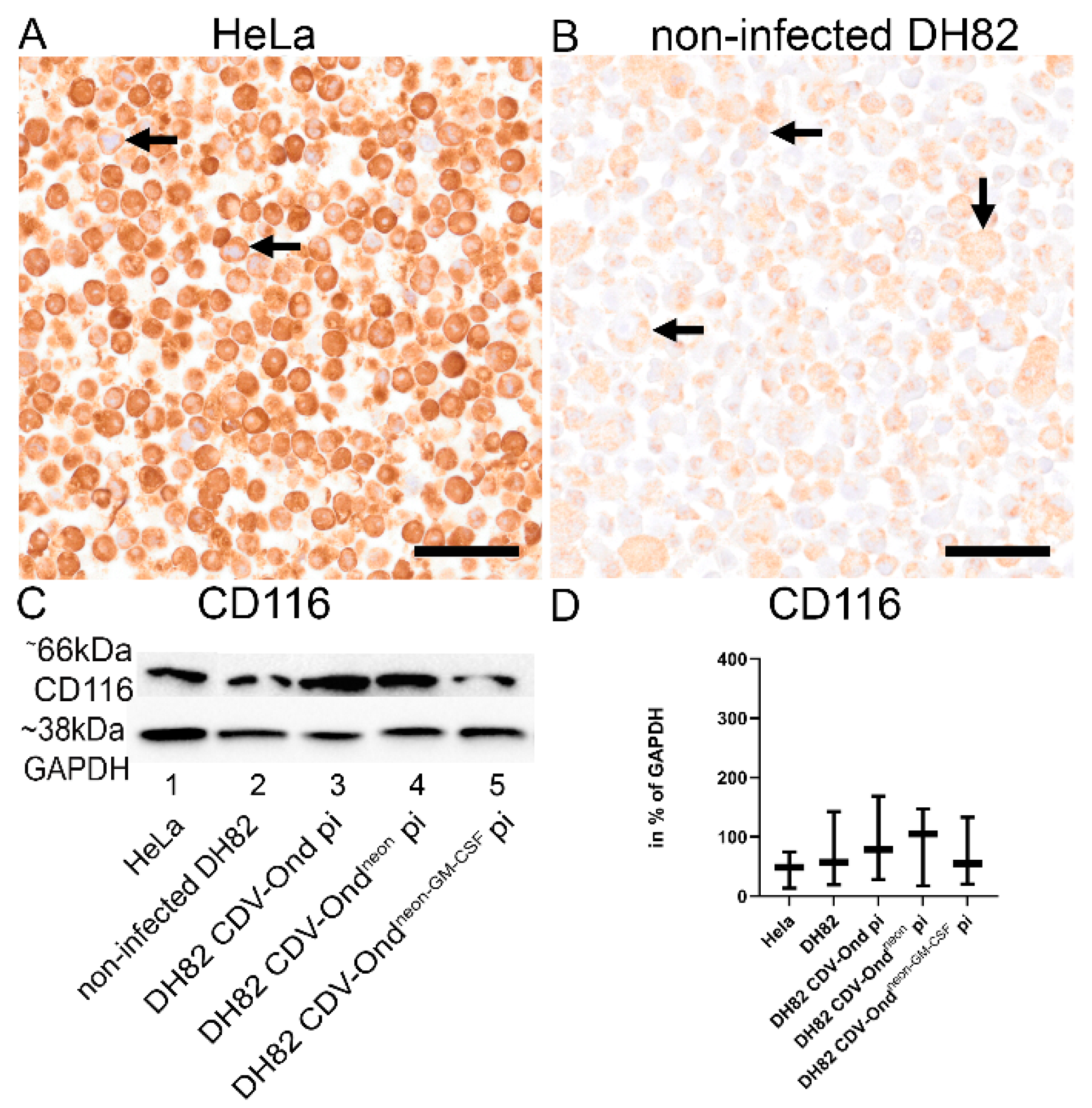
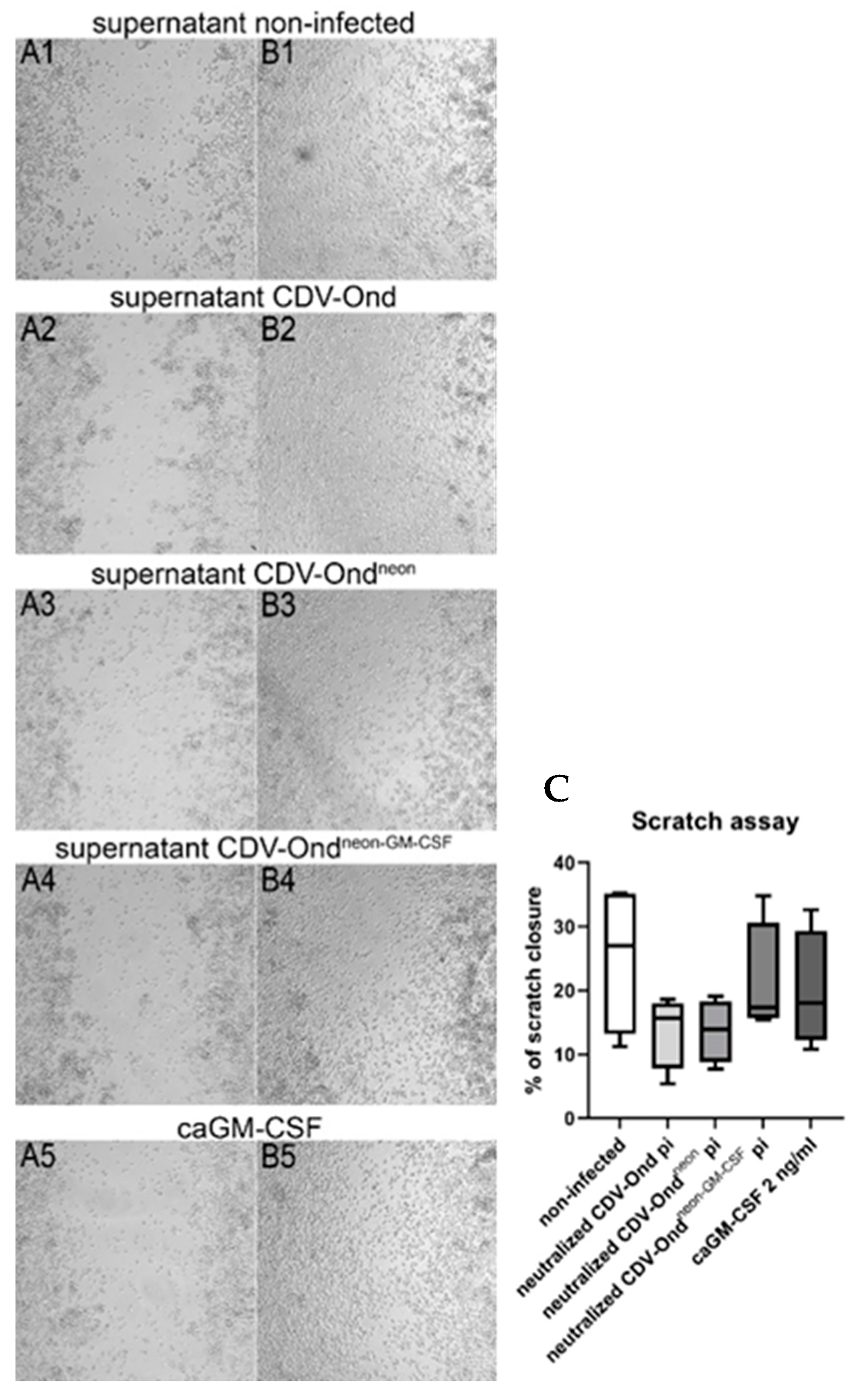
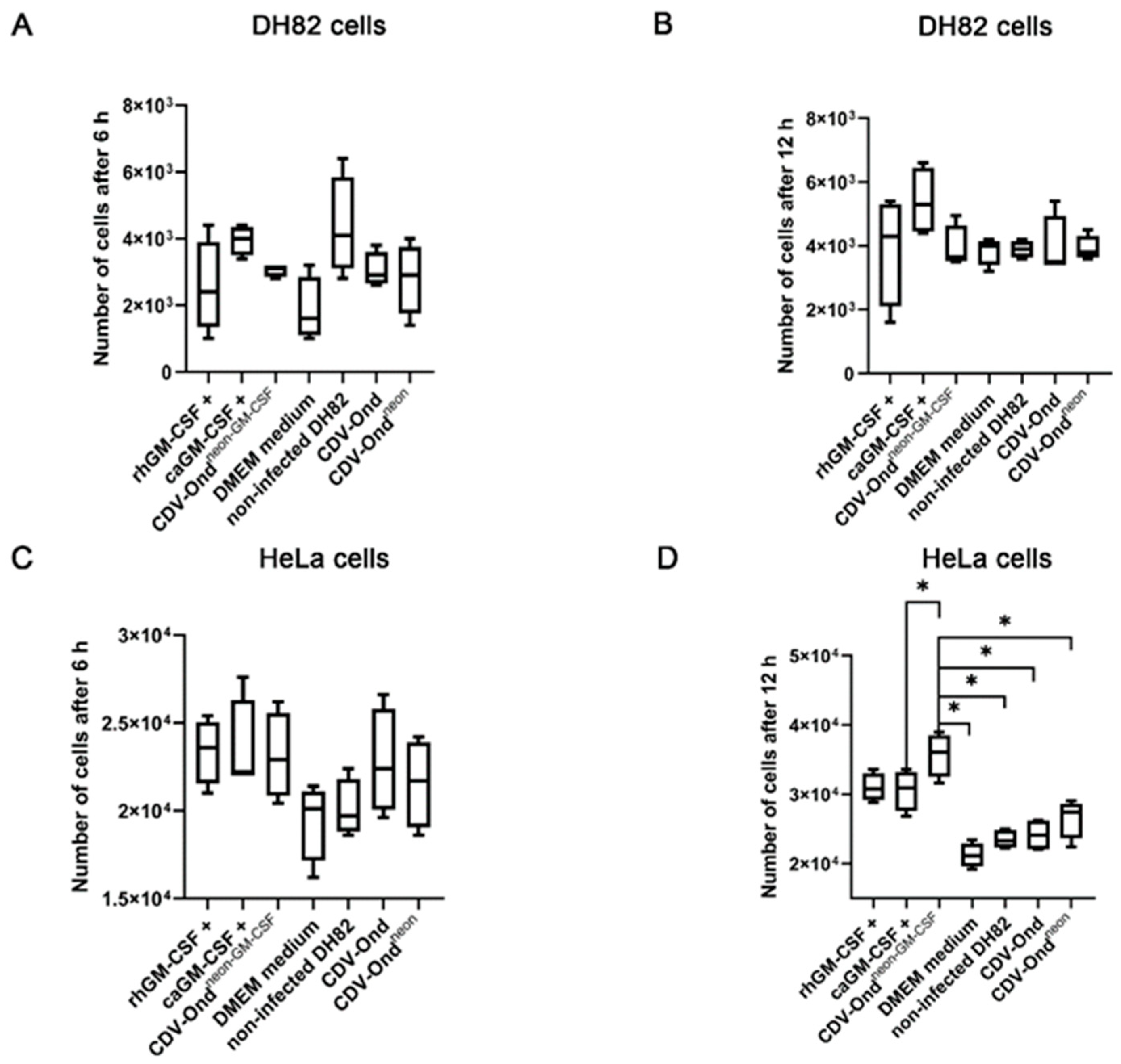
3.3. GM-CSF Produced by CDV-Ondneon-GM-CSF-Infected DH82 Cells Is Functional and Does Not Affect DH82 Cell Behavior
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dervisis, N.G.; Kiupel, M.; Qin, Q.; Cesario, L. Clinical prognostic factors in canine histiocytic sarcoma. Vet. Comp. Oncol. 2017, 15, 1171–1180. [Google Scholar] [CrossRef]
- Kennedy, K.; Thomas, R.; Breen, M. Canine histiocytic malignancies—Challenges and opportunities. Vet. Sci. 2016, 3, 2. [Google Scholar] [CrossRef]
- Affolter, V.K.; Moore, P.F. Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet. Pathol. 2002, 39, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Erich, S.A.; Rutteman, G.R.; Teske, E. Causes of death and the impact of histiocytic sarcoma on the life expectancy of the Dutch population of Bernese mountain dogs and Flat-coated retrievers. Vet. J. 2013, 198, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Moore, P.F. A review of histiocytic diseases of dogs and cats. Vet. Pathol. 2014, 51, 167–184. [Google Scholar] [CrossRef]
- Kadowaki, N. Intratumoral cancer immunotherapy exploiting anti-viral immunity. J. Clin. Exp. Hematop. 2022, 62, 1–8. [Google Scholar] [CrossRef]
- Melcher, A.; Harrington, K.; Vile, R. Oncolytic virotherapy as immunotherapy. Science 2021, 374, 1325–1326. [Google Scholar] [CrossRef] [PubMed]
- Howells, A.; Marelli, G.; Lemoine, N.R.; Wang, Y. Oncolytic viruses-interaction of virus and tumor cells in the battle to eliminate cancer. Front. Oncol. 2017, 7, 195. [Google Scholar] [CrossRef]
- Grote, D.; Russell, S.J.; Cornu, T.I.; Cattaneo, R.; Vile, R.; Poland, G.A.; Fielding, A.K. Live attenuated measles virus induces regression of human lymphoma xenografts in immunodeficient mice. Blood 2001, 97, 3746–3754. [Google Scholar] [CrossRef]
- Marek, K.; Armando, F.; Nippold, V.M.; Rohn, K.; Plattet, P.; Brogden, G.; Gerold, G.; Baumgärtner, W.; Puff, C. Persistent infection of a canine histiocytic sarcoma cell line with attenuated canine distemper virus expressing vasostatin or granulocyte-macrophage colony-stimulating factor. Int. J. Mol. Sci. 2022, 23, 6156. [Google Scholar] [CrossRef]
- Grossardt, C.; Engeland, C.E.; Bossow, S.; Halama, N.; Zaoui, K.; Leber, M.F.; Springfeld, C.; Jaeger, D.; von Kalle, C.; Ungerechts, G. Granulocyte-macrophage colony-stimulating factor-armed oncolytic measles virus is an effective therapeutic cancer vaccine. Hum. Gene Ther. 2013, 24, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Baird, S.M.; Powell, H.C.; Sharma, S.; Eling, D.J.; Kipps, T.J. Local and systemic effects after adenoviral transfer of the murine granulocyte-macrophage colony-stimulating factor gene into mice. Br. J. Haematol. 2000, 108, 641–652. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, H.M.; Ragsdale, C.E.; Gale, R.P.; Lyman, G.H. Sargramostim (rhu GM-CSF) as cancer therapy (Systematic review) and an immunomodulator. A drug before its time? Front. Immunol. 2021, 12, 706186. [Google Scholar] [CrossRef] [PubMed]
- Zarei, S.; Schwenter, F.; Luy, P.; Aurrand-Lions, M.; Morel, P.; Kopf, M.; Dranoff, G.; Mach, N. Role of GM-CSF signaling in cell-based tumor immunization. Blood 2009, 113, 6658–6668. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Ruby, C.E.; Hughes, T.; Slingluff, C.L., Jr. Current status of granulocyte-macrophage colony-stimulating factor in the immunotherapy of melanoma. J. Immunother. Cancer 2014, 2, 11. [Google Scholar] [CrossRef]
- Kumar, A.; Taghi Khani, A.; Sanchez Ortiz, A.; Swaminathan, S. GM-CSF: A double-edged sword in cancer immunotherapy. Front. Immunol. 2022, 13, 901277. [Google Scholar] [CrossRef]
- Salva, E.; Turan, S.O.; Akbuğa, J. Increased in vitro cell proliferation by chitosan/pGM-CSF complexes. Indian J. Pharm. Sci. 2011, 73, 131–138. [Google Scholar] [CrossRef]
- Russell, L.; Peng, K.W. The emerging role of oncolytic virus therapy against cancer. Chin. Clin. Oncol. 2018, 7, 16. [Google Scholar] [CrossRef]
- Kumar, A.; Parshad, R.; Suhani; Bhattacharjee, H.K.; Sharma, R. Thoracolaparoscopic repair of diaphragmatic hernias. Indian J. Thorac. Cardiovasc. Surg. 2021, 37, 558–564. [Google Scholar] [CrossRef]
- Eubank, T.D.; Roberts, R.; Galloway, M.; Wang, Y.; Cohn, D.E.; Marsh, C.B. GM-CSF induces expression of soluble VEGF receptor-1 from human monocytes and inhibits angiogenesis in mice. Immunity 2004, 21, 831–842. [Google Scholar] [CrossRef]
- Hong, I.S. Stimulatory versus suppressive effects of GM-CSF on tumor progression in multiple cancer types. Exp. Mol. Med. 2016, 48, e242. [Google Scholar] [CrossRef] [PubMed]
- Grote, D.; Cattaneo, R.; Fielding, A.K. Neutrophils contribute to the measles virus-induced antitumor effect: Enhancement by granulocyte macrophage colony-stimulating factor expression. Cancer Res. 2003, 63, 6463–6468. [Google Scholar] [PubMed]
- Yu, T.W.; Chueh, H.Y.; Tsai, C.C.; Lin, C.T.; Qiu, J.T. Novel GM-CSF-based vaccines: One small step in GM-CSF gene optimization, one giant leap for human vaccines. Hum. Vaccines Immunother. 2016, 12, 3020–3028. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, F.; Lehmbecker, A.; Raddatz, B.B.; Kegler, K.; Tipold, A.; Stein, V.M.; Kalkuhl, A.; Deschl, U.; Baumgärtner, W.; Ulrich, R.; et al. Morphologic, phenotypic, and transcriptomic characterization of classically and alternatively activated canine blood-derived macrophages in vitro. PLoS ONE 2017, 12, e0183572. [Google Scholar] [CrossRef]
- Bristol, J.A.; Zhu, M.; Ji, H.; Mina, M.; Xie, Y.; Clarke, L.; Forry-Schaudies, S.; Ennist, D.L. In vitro and in vivo activities of an oncolytic adenoviral vector designed to express GM-CSF. Mol. Ther. 2003, 7, 755–764. [Google Scholar] [CrossRef]
- Eubank, T.D.; Roberts, R.D.; Khan, M.; Curry, J.M.; Nuovo, G.J.; Kuppusamy, P.; Marsh, C.B. Granulocyte macrophage colony-stimulating factor inhibits breast cancer growth and metastasis by invoking an anti-angiogenic program in tumor-educated macrophages. Cancer Res. 2009, 69, 2133–2140. [Google Scholar] [CrossRef] [PubMed]
- Soiffer, R.; Lynch, T.; Mihm, M.; Jung, K.; Rhuda, C.; Schmollinger, J.C.; Hodi, F.S.; Liebster, L.; Lam, P.; Mentzer, S.; et al. Vaccination with irradiated autologous melanoma cells engineered to secrete human granulocyte-macrophage colony-stimulating factor generates potent antitumor immunity in patients with metastatic melanoma. Proc. Natl. Acad. Sci. USA 1998, 95, 13141–13146. [Google Scholar] [CrossRef]
- Mueller, M.M.; Fusenig, N.E. Constitutive expression of G-CSF and GM-CSF in human skin carcinoma cells with functional consequence for tumor progression. Int. J. Cancer 1999, 83, 780–789. [Google Scholar] [CrossRef]
- Armando, F.; Gambini, M.; Corradi, A.; Becker, K.; Marek, K.; Pfankuche, V.M.; Mergani, A.E.; Brogden, G.; de Buhr, N.; von Köckritz-Blickwede, M.; et al. Mesenchymal to epithelial transition driven by canine distemper virus infection of canine histiocytic sarcoma cells contributes to a reduced cell motility in vitro. J. Cell Mol. Med. 2020, 7, 9332–9348. [Google Scholar] [CrossRef]
- Armando, F.; Gambini, M.; Corradi, A.; Giudice, C.; Pfankuche, V.M.; Brogden, G.; Attig, F.; von Köckritz-Blickwede, M.; Baumgärtner, W.; Puff, C. Oxidative stress in canine histiocytic sarcoma cells induced by an infection with canine distemper virus led to a dysregulation of HIF-1alpha downstream pathway resulting in a reduced expression of VEGF-B in vitro. Viruses 2020, 12, 200. [Google Scholar] [CrossRef]
- Fayyad, A.; Lapp, S.; Risha, E.; Pfankuche, V.M.; Rohn, K.; Barthel, Y.; Schaudien, D.; Baumgärtner, W.; Puff, C. Matrix metalloproteinases expression in spontaneous canine histiocytic sarcomas and its xenograft model. Vet. Immunol. Immunopathol. 2018, 198, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Pfankuche, V.M.; Sayed-Ahmed, M.; Contioso, V.B.; Spitzbarth, I.; Rohn, K.; Ulrich, R.; Deschl, U.; Kalkuhl, A.; Baumgärtner, W.; Puff, C. Persistent morbillivirus infection leads to altered cortactin distribution in histiocytic sarcoma cells with decreased cellular migration capacity. PLoS ONE 2016, 11, e0167517. [Google Scholar] [CrossRef] [PubMed]
- Pfankuche, V.M.; Spitzbarth, I.; Lapp, S.; Ulrich, R.; Deschl, U.; Kalkuhl, A.; Baumgärtner, W.; Puff, C. Reduced angiogenic gene expression in morbillivirus-triggered oncolysis in a translational model for histiocytic sarcoma. J. Cell Mol. Med. 2017, 21, 816–830. [Google Scholar] [CrossRef] [PubMed]
- Puff, C.; Krudewig, C.; Imbschweiler, I.; Baumgärtner, W.; Alldinger, S. Influence of persistent canine distemper virus infection on expression of RECK, matrix-metalloproteinases and their inhibitors in a canine macrophage/monocytic tumour cell line (DH82). Vet. J. 2009, 182, 100–107. [Google Scholar] [CrossRef]
- Armando, F.; Fayyad, A.; Arms, S.; Barthel, Y.; Schaudien, D.; Rohn, K.; Gambini, M.; Lombardo, M.S.; Beineke, A.; Baumgärtner, W.; et al. Intratumoral canine distemper virus infection inhibits tumor growth by modulation of the tumor microenvironment in a murine xenograft model of canine histiocytic sarcoma. Int. J. Mol. Sci. 2021, 22, 3578. [Google Scholar] [CrossRef]
- Wyss, M.; Gradauskaite, V.; Ebert, N.; Thiel, V.; Zürbriggen, A.; Plattet, P. Efficient recovery of attenuated canine distemper virus from cDNA. Virus Res. 2022, 316, 198796. [Google Scholar] [CrossRef]
- Techangamsuwan, S.; Haas, L.; Rohn, K.; Baumgärtner, W.; Wewetzer, K. Distinct cell tropism of canine distemper virus strains to adult olfactory ensheathing cells and Schwann cells in vitro. Virus Res. 2009, 144, 195–201. [Google Scholar] [CrossRef]
- Frisk, A.L.; König, M.; Moritz, A.; Baumgärtner, W. Detection of canine distemper virus nucleoprotein RNA by reverse transcription-PCR using serum, whole blood, and cerebrospinal fluid from dogs with distemper. J. Clin. Microbiol. 1999, 37, 3634–3643. [Google Scholar] [CrossRef]
- Armando, F.; Ferrari, L.; Arcari, M.L.; Azzali, G.; Dallatana, D.; Ferrari, M.; Lombardi, G.; Zanfabro, M.; Di Lecce, R.; Lunghi, P.; et al. Endocanalicular transendothelial crossing (ETC): A novel intravasation mode used by HEK-EBNA293-VEGF-D cells during the metastatic process in a xenograft model. PLoS ONE 2020, 15, e0239932. [Google Scholar] [CrossRef]
- Santos, A.X.; Maia, J.E.; Crespo, P.M.; Pettenuzzo, L.F.; Daniotti, J.L.; Barbé-Tuana, F.M.; Martins, L.M.; Trindade, V.M.; Borojevic, R.; Guma, F.C. GD1a modulates GM-CSF-induced cell proliferation. Cytokine 2011, 56, 600–607. [Google Scholar] [CrossRef]
- Doka, R.M.; Suter, S.E.; Mastromauro, M.L.; Bennett, A.L.; Hess, P.R. Doxorubicin for treatment of histiocytic sarcoma in dogs: 31 cases (2003–2017). J. Am. Vet. Med. Assoc. 2022, 260, 1827–1833. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J. Lomustine chemotherapy for the treatment of presumptive haemophagocytic histiocytic sarcoma in Flat-coated Retrievers. Aust. Vet. J. 2018, 96, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Schwens, C.; Thom, N.; Moritz, A. Reactive and neoplastic histiocytic diseases in the dog. Tierarztl. Prax. 2011, 39, 176–190. [Google Scholar]
- Bindrich, H. Beitrag zum Wesen der Staupevirusinfektion des Hundes und zu ihrer Bekämpfung. Arch. Exp. Vet. Med. 1954, 8, 263–315. [Google Scholar]
- Wettreich, A.; Sebollela, A.; Carvalho, M.A.; Azevedo, S.P.; Borojevic, R.; Ferreira, S.T.; Coelho-Sampaio, T. Acidic pH modulates the interaction between human granulocyte-macrophage colony-stimulating factor and glycosaminoglycans. J. Biol. Chem. 1999, 274, 31468–31475. [Google Scholar] [CrossRef]
- Kolvenbach, C.G.; Narhi, L.O.; Philo, J.S.; Li, T.; Zhang, M.; Arakawa, T. Granulocyte-colony stimulating factor maintains a thermally stable, compact, partially folded structure at pH2. J. Pept. Res. 1997, 50, 310–318. [Google Scholar] [CrossRef]
- Rivas, C.I.; Vera, J.C.; Delgado-López, F.; Heaney, M.L.; Guaiquil, V.H.; Zhang, R.H.; Scher, H.I.; Concha, I.I.; Nualart, F.; Cordon-Cardo, C.; et al. Expression of granulocyte-macrophage colony-stimulating factor receptors in human prostate cancer. Blood 1998, 91, 1037–1043. [Google Scholar] [CrossRef]
- Guthridge, M.A.; Powell, J.A.; Barry, E.F.; Stomski, F.C.; McClure, B.J.; Ramshaw, H.; Felquer, F.A.; Dottore, M.; Thomas, D.T.; To, B.; et al. Growth factor pleiotropy is controlled by a receptor Tyr/Ser motif that acts as a binary switch. EMBO J. 2006, 25, 479–489. [Google Scholar] [CrossRef]
- Zhan, Y.; Lew, A.M.; Chopin, M. The pleiotropic effects of the GM-CSF Rheostat on myeloid cell differentiation and function: More than a numbers game. Front. Immunol. 2019, 10, 2679. [Google Scholar] [CrossRef]


| Primary Antibody | Host Species, Clonality | Blocking Serum | Dilution | Secondary Antibody (1:200) |
|---|---|---|---|---|
| CDV-NP (University of Bern, Prof. Zurbriggen) | Mouse, monoclonal, clone D110 | Goat serum | 1:100 (IF) | GaM-Cy3 |
| CD116 (Invitrogen, CA, USA) | Rabbit, polyclonal | Goat serum | 1:500 (IHC) | GaR-b |
| Gene | Primer Sequence (5′-3′) | Amplicon Length (bp) | Position | GenBank Accession Number | |
|---|---|---|---|---|---|
| CDV # | Forward * | ACAGGATTGCTGAGGACCTAT | 287 | 769–789 | AF378705 |
| Reverse * | CAAGATAACCATGTACGGTGC | 1055–1035 | |||
| Forward | GCTCTTGGGTTGCATGAGTT | 83 | 954–973 | ||
| Reverse | GCTGTTTCACCCATCTGTTG | 1036–1017 | |||
| TCID50/mL | ||
|---|---|---|
| Range w/o Acidification | Range After Acidification | |
| non-infected control | - | - |
| CDV-Ond pi | 103–104 | - |
| CDV-Ondneon pi | 103–103.75 | - |
| CDV-Ondneon-GM-CSF pi | 103–104 | - |
| Infection Status | Number of CDV mRNA Transcripts in Supernatants without Acidification Median (Range) | Number of CDV mRNA Transcripts in Supernatants after Acidification Median (Range) |
|---|---|---|
| Non-infected | 0 (0.00–0.00) | 0 (0.00–0.00) |
| CDV-Ond pi | 2841.86 (668.86–10154.25) | 156.98 (93.92–495.60) |
| CDV-Ondneon pi | 4287.86 (1132.87–17252.94) | 122.14 (103.97–237.24) |
| CDV-Ondneon-GM-CSF pi | 17508.73 (6657.71–87199.45) | 821.93 (514.62–1481.72) |
| Infection Status | CDV nucleoprotein IOD in Supernatants without Acidification Median (Range) | CDV nucleoprotein IOD in Supernatants after Acidification Median (Range) |
|---|---|---|
| Non-infected | 0.00 (0.00–0.00) | 0.00 (0.00–0.00) |
| CDV-Ond pi | 11.05 (4.12–33.32) | 13.24 (2.96–13.44) |
| CDV-Ondneon pi | 58.11 (45.88–112.42) | 45.24 (42.21–124.92) |
| CDV-Ondneon-GM-CSF pi | 113.84 (84.48–155.00) | 142.68 (99.00–154.71) |
| Infection Status | GM-CSF IOD in Supernatants without Acidification Median (Range) | GM-CSF IOD in Supernatants after Acidification Median (Range) |
|---|---|---|
| Non-infected | 0.00 (0.00–0.00) | 0.00 (0.00–0.00) |
| CDV-Ond pi | 0.00 (0.00–0.00) | 0.00 (0.00–0.00) |
| CDV-Ondneon pi | 0.00 (0.00–0.00) | 0.00 (0.00–0.00) |
| CDV-Ondneon-GM-CSF pi | 130.48 (62.98–150.29) | 131.08 (45.29–153.36) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marek, K.; Armando, F.; Asawapattanakul, T.; Nippold, V.M.; Plattet, P.; Gerold, G.; Baumgärtner, W.; Puff, C. Functional Granulocyte–Macrophage Colony-Stimulating Factor (GM-CSF) Delivered by Canine Histiocytic Sarcoma Cells Persistently Infected with Engineered Attenuated Canine Distemper Virus. Pathogens 2023, 12, 877. https://doi.org/10.3390/pathogens12070877
Marek K, Armando F, Asawapattanakul T, Nippold VM, Plattet P, Gerold G, Baumgärtner W, Puff C. Functional Granulocyte–Macrophage Colony-Stimulating Factor (GM-CSF) Delivered by Canine Histiocytic Sarcoma Cells Persistently Infected with Engineered Attenuated Canine Distemper Virus. Pathogens. 2023; 12(7):877. https://doi.org/10.3390/pathogens12070877
Chicago/Turabian StyleMarek, Katarzyna, Federico Armando, Thanaporn Asawapattanakul, Vanessa Maria Nippold, Philippe Plattet, Gisa Gerold, Wolfgang Baumgärtner, and Christina Puff. 2023. "Functional Granulocyte–Macrophage Colony-Stimulating Factor (GM-CSF) Delivered by Canine Histiocytic Sarcoma Cells Persistently Infected with Engineered Attenuated Canine Distemper Virus" Pathogens 12, no. 7: 877. https://doi.org/10.3390/pathogens12070877
APA StyleMarek, K., Armando, F., Asawapattanakul, T., Nippold, V. M., Plattet, P., Gerold, G., Baumgärtner, W., & Puff, C. (2023). Functional Granulocyte–Macrophage Colony-Stimulating Factor (GM-CSF) Delivered by Canine Histiocytic Sarcoma Cells Persistently Infected with Engineered Attenuated Canine Distemper Virus. Pathogens, 12(7), 877. https://doi.org/10.3390/pathogens12070877







