Characterization of an Escherichia coli Isolate Coharboring the Virulence Gene astA and Tigecycline Resistance Gene tet(X4) from a Dead Piglet
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pathogen Detection
2.2. Bacterial Isolation and Identification
2.3. Antimicrobial Susceptibility Testing
2.4. Whole-Genome Sequencing (WGS) and Bioinformatics Analysis
2.5. Conjugative Transfer
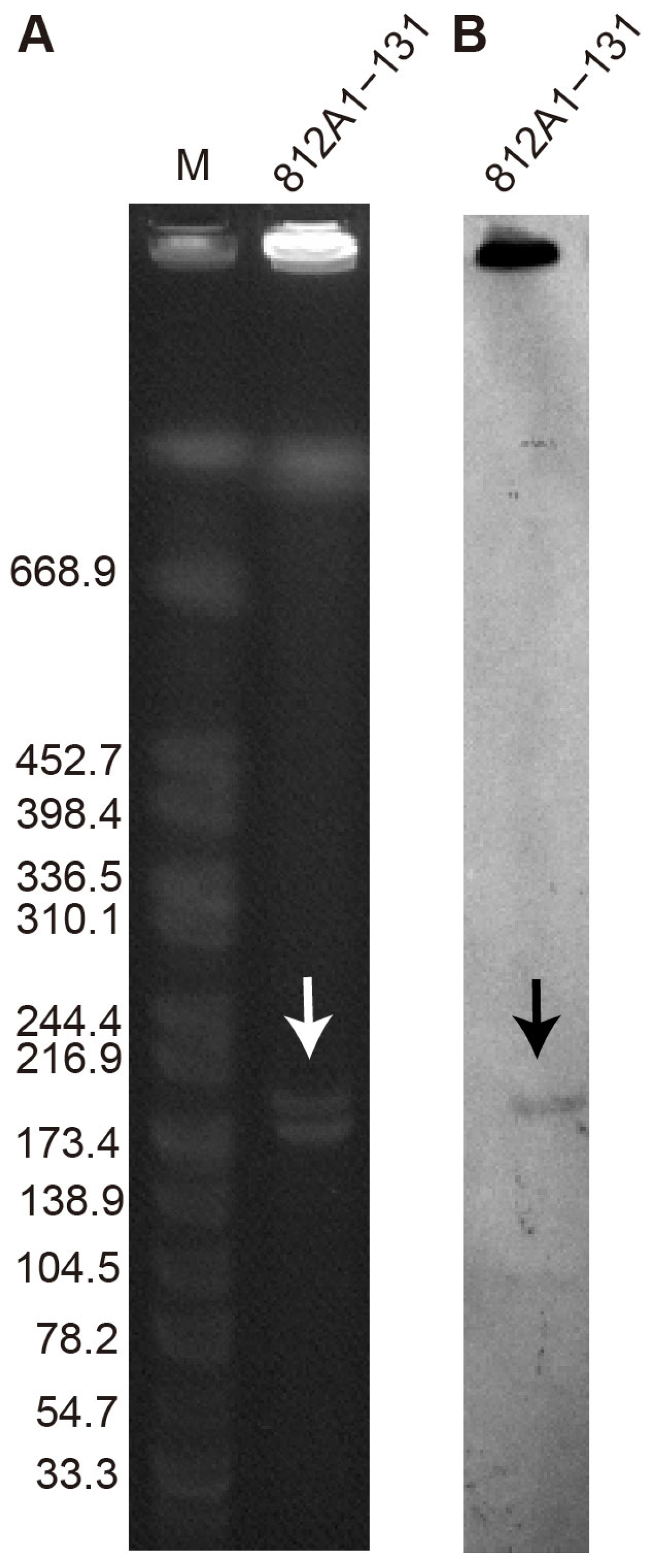
2.6. S1-PFGE and Southern Blotting
2.7. The Galleria mellonella Model and the Mouse Infection Model
3. Results
3.1. Pathogen Detection and Bacterial Isolation
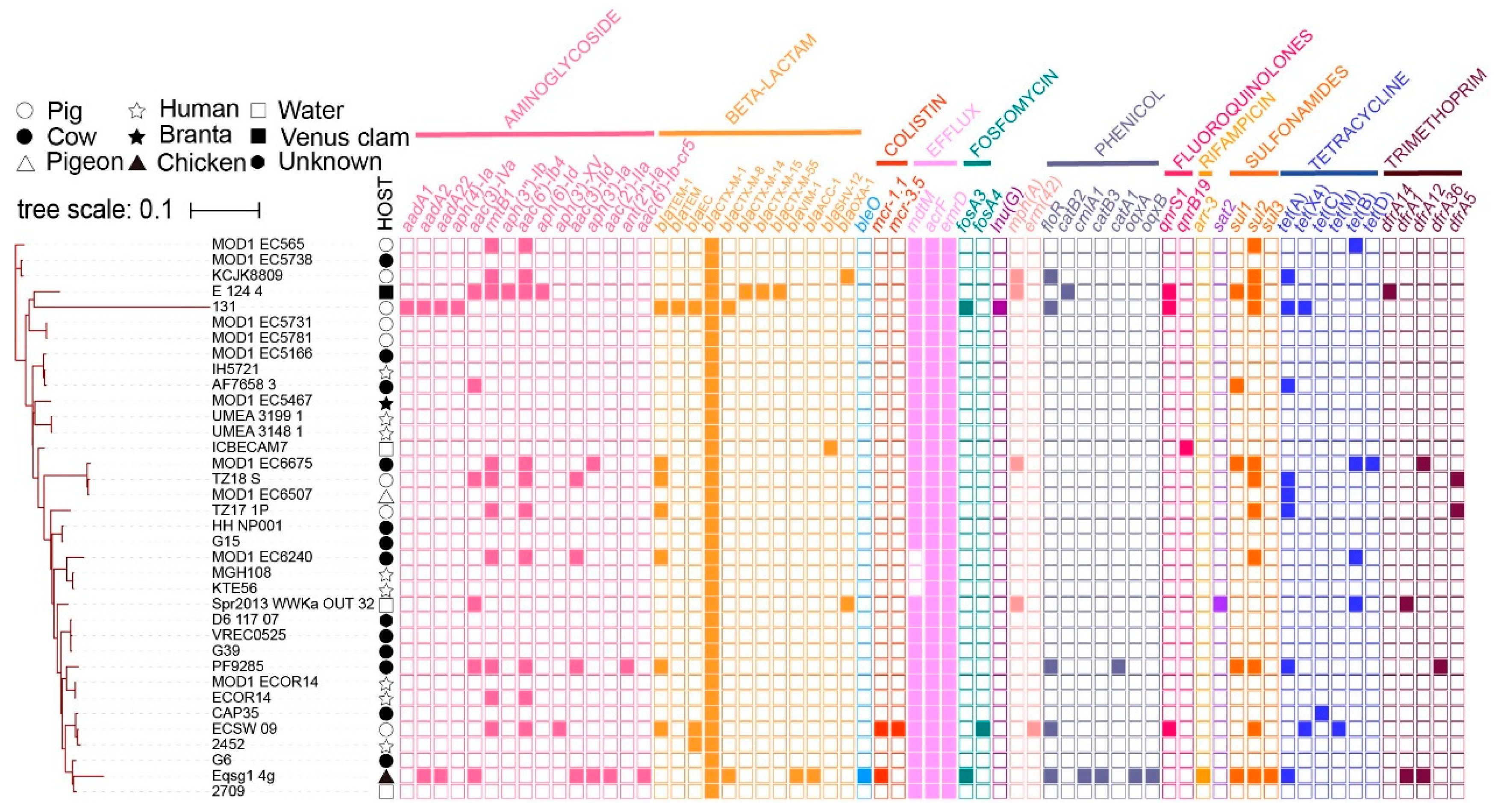
3.2. Antimicrobial Susceptibility
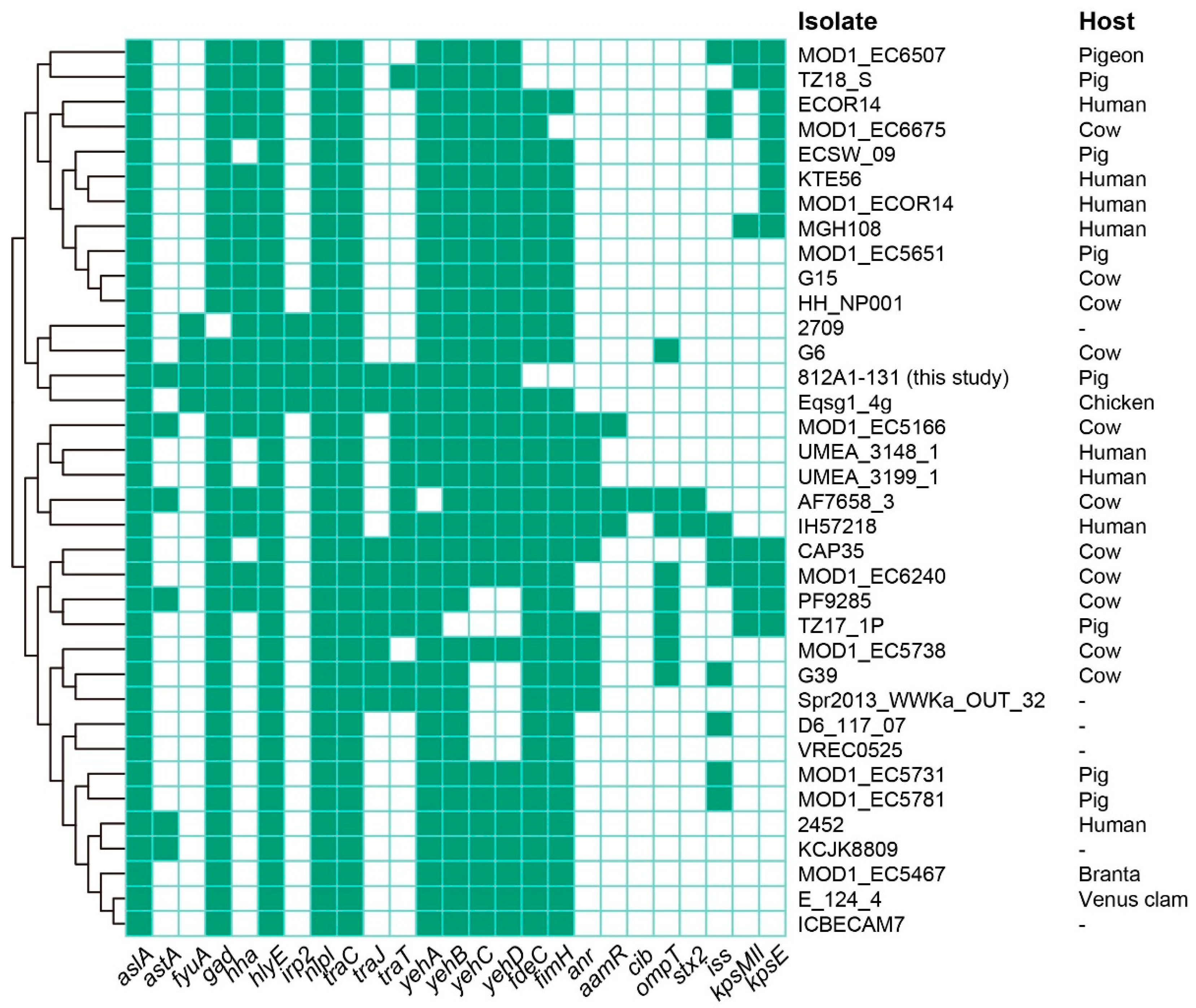
3.3. Whole-Genome Sequencing and Sequence Analysis
3.4. Conjugative Transfer of 812A1-131
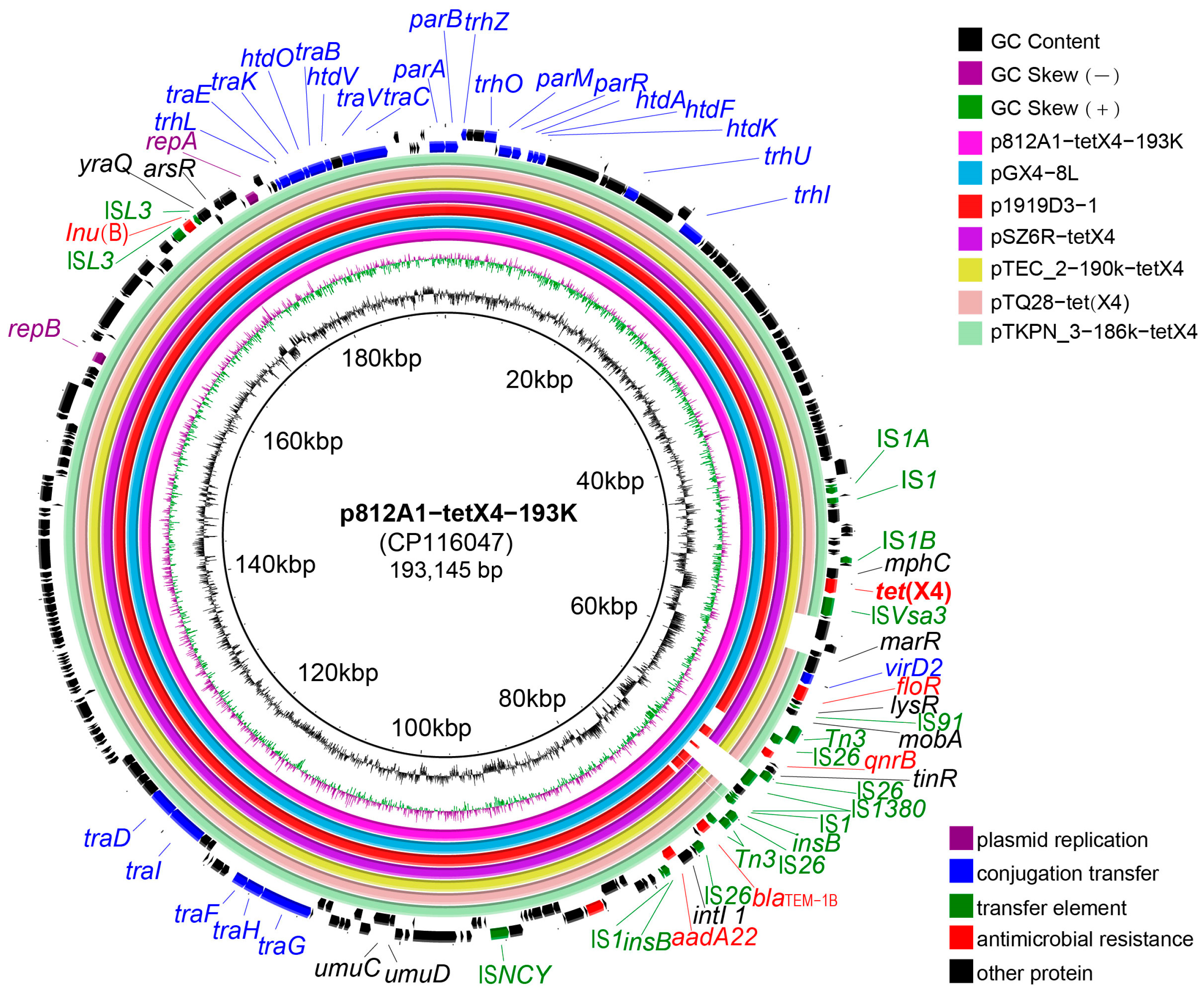
3.5. Genetic Environment Analysis of p812A1-tetX4-193K
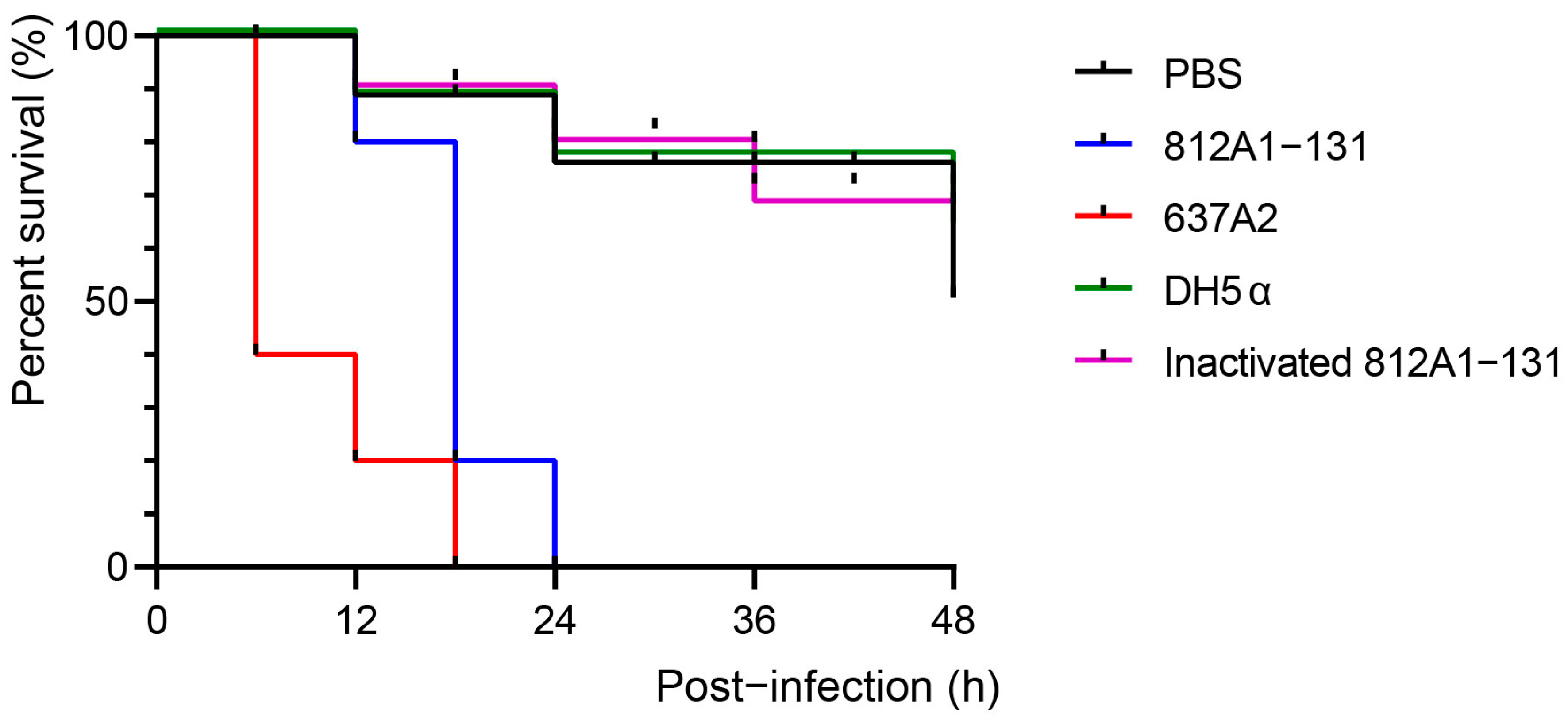
3.6. Pathogenic Testing of Strain 812A1-131 in Galleria mellonella and Mouse
4. Discussion
5. Accession Numbers
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z.; et al. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Frey, L.; Tanunchai, B.; Glaser, B. Antibiotics residues in pig slurry and manure and its environmental contamination potential. A meta-analysis. Agron. Sustain. Dev. 2022, 42, 31. [Google Scholar] [CrossRef]
- Zhang, R.; Dong, N.; Shen, Z.; Zeng, Y.; Lu, J.; Liu, C.; Zhou, H.; Hu, Y.; Sun, Q.; Cheng, Q. Epidemiological and phylogenetic analysis reveals Flavobacteriaceae as potential ancestral source of tigecycline resistance gene tet(X). Nat. Commun. 2020, 11, 4648. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.Y.; Li, X.J.; Chen, C.; Wu, X.T.; He, Q.; Jia, Q.L.; Zhang, X.J.; Lin, Z.Y.; Li, C.; Fang, L.X.; et al. Comprehensive analysis of plasmid-mediated tet(X4)-positive Escherichia coli isolates from clinical settings revealed a high correlation with animals and environments-derived strains. Sci. Total Environ. 2022, 806, 150687. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Tang, B.; Yang, H.; Ma, J.; Huang, Y.; Liu, C. Emergence of plasmid-mediated tigecycline resistance gene, tet(X4), in Escherichia fergusonii from pigs. J. Glob. Antimicrob. Resist. 2022, 30, 249–251. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Tian, Y.; Shao, D.; Zhang, M.; Li, J.; Song, H.; Sun, C.; Wang, Y.; Liu, D.; Zhang, Y. Fecal carriage of Escherichia coli harboring the tet(X4)-IncX1 plasmid from a tertiary class-a hospital in Beijing, China. Antibiotics 2022, 11, 1068. [Google Scholar] [CrossRef]
- Li, R.; Liu, Z.; Li, Y.; Xiao, X.; Wang, Z. Characterization of blaNDM-positive Enterobacteriaceae reveals the clonal dissemination of Enterobacter hormaechei coharboring blaNDM and tet(X4) along the pork production chain. Int. J. Food Microbiol. 2022, 372, 109692. [Google Scholar] [CrossRef]
- Li, R.; Lu, X.; Munir, A.; Abdullah, S.; Liu, Y.; Xiao, X.; Wang, Z.; Mohsin, M. Widespread prevalence and molecular epidemiology of tet(X4) and mcr-1 harboring Escherichia coli isolated from chickens in Pakistan. Sci. Total Environ. 2022, 806, 150689. [Google Scholar] [CrossRef]
- Ma, J.; Zhou, W.; Wu, J.; Liu, X.; Lin, J.; Ji, X.; Lin, H.; Wang, J.; Jiang, H.; Zhou, Q.; et al. Large-scale studies on antimicrobial resistance and molecular characterization of Escherichia coli from food animals in developed areas of Eastern China. Microbiol. Spectr. 2022, 10, e0201522. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Cui, C.Y.; Zhang, Y.; He, Q.; Wu, X.T.; Li, G.; Liao, X.P.; Kreiswirth, B.N.; Liu, Y.H.; Chen, L.; et al. Emergence of mobile tigecycline resistance mechanism in Escherichia coli strains from migratory birds in China. Emerg. Microbes Infect. 2019, 8, 1219–1222. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Wan, Y.; Du, P.; Liu, D.; Li, R.; Zhang, P.; Wu, Y.; Fanning, S.; Wang, Y.; Bai, L. Investigation of tigecycline resistant Escherichia coli from raw meat reveals potential transmission among food-producing animals. Food Control 2021, 121, 107633. [Google Scholar] [CrossRef]
- Li, R.; Mohsin, M.; Lu, X.; Abdullah, S.; Munir, A.; Wang, Z. Emergence of plasmid-mediated resistance genes tet(X) and mcr-1 in Escherichia coli clinical isolates from Pakistan. mSphere 2021, 6, e0069521. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, K.J.; Roberts, J.L.; Collignon, P. Escherichia coli bacteraemia in Canberra: Incidence and clinical features. Med. J. Aust. 2008, 188, 209–213. [Google Scholar] [CrossRef]
- Yayan, J.; Ghebremedhin, B.; Rasche, K. No Development of imipenem resistance in pneumonia caused by Escherichia coli. Medicine 2015, 94, e1020. [Google Scholar] [CrossRef]
- Bradford, P.A.; Petersen, P.J.; Fingerman, I.M.; White, D.G. Characterization of expanded-spectrum cephalosporin resistance in E. coli isolates associated with bovine calf diarrhoeal disease. J. Antimicrob. Chemother. 1999, 44, 607–610. [Google Scholar] [CrossRef]
- Kemmett, K.; Williams, N.; Chaloner, G.; Humphrey, S.; Wigley, P.; Humphrey, T. The contribution of systemic Escherichia coli infection to the early mortalities of commercial broiler chickens. Avian Pathol. 2014, 43, 37–42. [Google Scholar] [CrossRef]
- Aminshahidi, M.; Arastehfar, A.; Pouladfar, G.; Arman, E.; Fani, F. Diarrheagenic Escherichia coli and Shigella with high rate of extended-spectrum Beta-lactamase production: Two predominant etiological agents of acute diarrhea in Shiraz, Iran. Microb. Drug Resist. 2017, 23, 1037–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angulo-Zamudio, U.A.; Gutiérrez-Jiménez, J.; Monroy-Higuera, L.; Flores-Villaseñor, H.; Leon-Sicairos, N.; Velazquez-Roman, J.; Vidal, J.E.; Tapia-Pastrana, G.; Canizalez-Roman, A. Non-diarrheagenic and diarrheagenic E. coli carrying supplementary virulence genes (SVG) are associated with diarrhea in children from Mexico. Microb. Pathog. 2021, 157, 104994. [Google Scholar] [CrossRef]
- Clements, A.; Young, J.C.; Constantinou, N.; Frankel, G. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012, 3, 71–87. [Google Scholar] [CrossRef] [Green Version]
- Zajacova, Z.S.; Konstantinova, L.; Alexa, P. Detection of virulence factors of Escherichia coli focused on prevalence of EAST1 toxin in stool of diarrheic and non-diarrheic piglets and presence of adhesion involving virulence factors in astA positive strains. Vet. Microbiol. 2012, 154, 369–375. [Google Scholar] [CrossRef]
- Awad, W.S.; El-Sayed, A.A.; Mohammed, F.F.; Bakry, N.M.; Abdou, N.-E.M.I.; Kamel, M.S. Molecular characterization of pathogenic Escherichia coli isolated from diarrheic and in-contact cattle and buffalo calves. Trop. Anim. Health Prod. 2020, 52, 3173–3185. [Google Scholar] [CrossRef]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 2015, 161, 980–988. [Google Scholar] [CrossRef]
- Henriot, C.P.; Celle, H.; Klaba, V.; Biguenet, A.; Miège, C.; Daval, A.; Amiotte-Suchet, P.; Beugnot, J.-C.; Karbowiak, T.; Bertrand, X. Effect of a karst system (France) on extended spectrum beta-lactamase (ESBL)-producing Escherichia coli. Water Res. 2023, 230, 119582. [Google Scholar] [CrossRef] [PubMed]
- Seenama, C.; Thamlikitkul, V.; Ratthawongjirakul, P. Multilocus sequence typing and blaESBL characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolated from healthy humans and swine in Northern Thailand. Infect. Drug Resist. 2019, 12, 2201–2214. [Google Scholar] [CrossRef] [Green Version]
- Dos Santos Alves, T.; Rosa, V.S.; da Silva Leite, D.; Guerra, S.T.; Joaquim, S.F.; Guimarães, F.F.; de Figueiredo Pantoja, J.C.; Lucheis, S.B.; Rall, V.L.M.; Hernandes, R.T. Genome-based characterization of multidrug-resistant Escherichia coli isolated from clinical bovine mastitis. Microbiol. Res. 2023, 80, 89. [Google Scholar] [CrossRef]
- Jonare, L.; Östlund, E.; Söderlund, R.; Hansson, I.; Aspán, A.; Jansson, D.S. Core genome multilocus sequence typing (cgMLST) confirms systemic spread of avian pathogenic Escherichia coli (APEC) in broilers with cellulitis. Veter. Microbiol. 2023, 282, 109755. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, P.; Du, P.; Zhang, X.; Wang, J.; Yang, Y.; Sun, H.; Wang, Z.; Cui, S.; Li, R. Prevalence and genomic characteristics of mcr-positive Escherichia coli strains isolated from humans, pigs, and foods in China. Microbiol. Spectr. 2023, 12, e04569-22. [Google Scholar] [CrossRef] [PubMed]
- García-Meniño, I.; García, V.; Mora, A.; Díaz-Jiménez, D.; Flament-Simon, S.C.; Alonso, M.P.; Blanco, J.E.; Blanco, M.; Blanco, J. Swine enteric colibacillosis in Spain: Pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 2018, 9, 2659. [Google Scholar] [CrossRef]
- Wen, F.; Yang, J.; Li, A.; Gong, Z.; Yang, L.; Cheng, Q.; Wang, C.; Zhao, M.; Yuan, S.; Chen, Y.; et al. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS ONE 2021, 16, e0253622. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Garcia-Toledo, L.; Fasulo, D.; Gladney, L.M.; Strockbine, N. Multiplex polymerase chain reaction for identification of Escherichia coli, Escherichia albertii and Escherichia fergusonii. J. Microbiol. Methods 2017, 140, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Tang, B.; Zheng, X.; Chang, J.; Ma, J.; He, Y.; Yang, H.; Wu, Y. Emergence of Incl2 plasmid-mediated colistin resistance in avian Escherichia fergusonii. FEMS Microbiol. Lett. 2022, 369, fnac016. [Google Scholar] [CrossRef]
- Tang, B.; Wang, J.; Zheng, X.; Chang, J.; Ma, J.; Wang, J.; Ji, X.; Yang, H.; Ding, B. Antimicrobial resistance surveillance of Escherichia coli from chickens in the Qinghai Plateau of China. Front. Microbiol. 2022, 13, 885132. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Yin, Y.; Li, Y.; Qin, S.; Liu, Y.; Yang, X.; Wang, Z.; Li, R. QitanTech Nanopore long-read sequencing enables rapid resolution of complete genomes of multi-drug resistant pathogens. Front. Microbiol. 2022, 13, 778659. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Tang, B.; Dai, R.; Zhang, B.; Chen, L.; Yang, H.; Zhao, G.; Ding, X. Identification of the streptothricin and tunicamycin biosynthetic gene clusters by genome mining in Streptomyces sp. strain fd1-xmd. Appl. Microbiol. Biotechnol. 2018, 102, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Lin, R.; Zhou, Z.; Ma, J.; Lin, H.; Zheng, X.; Wang, J.; Wu, J.; Dong, Y.; Jiang, H.; et al. Corrigendum: Antimicrobial resistance and genomic characterization of Escherichia coli from pigs and chickens in Zhejiang, China. Front. Microbiol. 2023, 14, 1102931. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Siddique, A.; Jia, C.; Ed-Dra, A.; Wu, J.; Lin, H.; Yue, M. Genome-based risk assessment for foodborne Salmonella enterica from food animals in China: A One Health perspective. Int. J. Food Microbiol. 2023, 390, 110120. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garcìa-Fernandez, A.; Larsen, M.; Lund, O.; Voldby Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids. Antimicrob using PlasmidFinder and plasmid multilocus sequence typing. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Tetzschner, A.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Zou, S.; Chen, H.; Yu, Y.; Ruan, Z. BacWGSTdb 2.0: A one-stop repository for bacterial whole-genome sequence typing and source tracking. Nucleic Acids Res. 2021, 49, D644–D650. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [Green Version]
- Jønsson, R.; Struve, C.; Jenssen, H.; Krogfelt, K.A. The wax moth Galleria mellonella as a novel model system to study Enteroaggregative Escherichia coli pathogenesis. Virulence 2017, 8, 1894–1899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, C.J.-Y.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohawk, K.L.; Melton-Celsa, A.R.; Zangari, T.; Carroll, E.E.; O’Brien, A.D. Pathogenesis of Escherichia coli O157:H7 strain 86-24 following oral infection of BALB/c mice with an intact commensal flora. Microb. Pathog. 2010, 48, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Feng, L.; Wang, F.; Wang, L. Enterohemorrhagic Escherichia coli senses low biotin status in the large intestine for colonization and infection. Nat. Commun. 2015, 6, 6592. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, F.; Xu, X.; Huang, H.; Lyu, N.; Ma, S.; Chen, L.; Mao, M.; Hu, Y.; Song, X. Detection of plasmid-mediated tigecycline resistance gene tet(X4) in a Salmonella enterica Serovar Llandoff isolate. Infect. Microbes Dis. 2021, 3, 198–204. [Google Scholar] [CrossRef]
- Zhai, W.; Tian, Y.; Lu, M.; Zhang, M.; Song, H.; Fu, Y.; Ma, T.; Sun, C.; Bai, L.; Wang, Y.; et al. Presence of mobile tigecycline resistance gene tet(X4) in Clinical Klebsiella pneumoniae. Microbiol. Spectr. 2022, 10, e0108121. [Google Scholar] [CrossRef]
- Nguyen, M.N.; Hoang, H.T.T.; Xavier, B.B.; Lammens, C.; Le, H.T.; Hoang, N.T.B.; Nguyen, S.T.; Pham, N.T.; Goossens, H.; Dang, A.D. Prospective One Health genetic surveillance in Vietnam identifies distinct blaCTX-M-harbouring Escherichia coli in food-chain and human-derived samples. Clin. Microbiol. Infect. 2021, 27, 1515.e1–1515.e8. [Google Scholar] [CrossRef] [PubMed]
- García, V.; García-Meniño, I.; Mora, A.; Flament-Simon, S.C.; Díaz-Jiménez, D.; Blanco, J.E.; Alonso, M.P.; Blanco, J. Co-occurrence of mcr-1, mcr-4 and mcr-5 genes in multidrug-resistant ST10 Enterotoxigenic and Shiga toxin-producing Escherichia coli in Spain (2006–2017). Int. J. Antimicrob. Agents 2018, 52, 104–108. [Google Scholar] [CrossRef]
- Day, M.J.; Hopkins, K.L.; Wareham, D.W.; Toleman, M.A.; Elviss, N.; Randall, L.; Teale, C.; Cleary, P.; Wiuff, C.; Doumith, M. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: An epidemiological surveillance and typing study. Lancet Infect. Dis. 2019, 19, 1325–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madec, J.-Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-spectrum β-lactamase/AmpC-and carbapenemase-producing Enterobacteriaceae in animals: A threat for humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Fontana, H.; Sano, E.; Li, R.; Humayon, M.; Rahman, S.; Lincopan, N.; Mohsin, M. Genomic features of a high-risk mcr-1.1-positive Escherichia coli ST10 isolated from cattle farm environment. Environ. Sci. Pollut. Res. 2021, 28, 54147–54152. [Google Scholar] [CrossRef]
- Dos Anjos, C.; Sabino, C.P.; Bueris, V.; Fernandes, M.R.; Pogliani, F.C.; Lincopan, N.; Sellera, F.P. Antimicrobial blue light inactivation of international clones of multidrug-resistant Escherichia coli ST10, ST131 and ST648. Photodiagnosis Photodyn. Ther. 2019, 27, 51–53. [Google Scholar] [CrossRef]
- Ćwiek, K.; Woźniak-Biel, A.; Karwańska, M.; Siedlecka, M.; Lammens, C.; Rebelo, A.R.; Hendriksen, R.S.; Kuczkowski, M.; Chmielewska-Władyka, M.; Wieliczko, A. Phenotypic and genotypic characterization of mcr-1-positive multidrug-resistant Escherichia coli ST93, ST117, ST156, ST10, and ST744 isolated from poultry in Poland. Braz. J. Microbiol. 2021, 52, 1597–1609. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, C.; Zhang, J.; Hua, W.; He, R.; Zhou, M. Carbapenemase- and colistin resistant Escherichia coli strains from children in China: High genetic diversity and first report of bla(NDM-5), bla(CTX-M-65), bla(OXA-10), bla(TEM-1), and mcr-1.1 genes co-occurrence in E. coli ST156. Infect. Drug Resist. 2022, 15, 5315–5320. [Google Scholar] [CrossRef]
- Lekunberri, I.; Balcázar, J.L.; Borrego, C.M. Metagenomic exploration reveals a marked change in the river resistome and mobilome after treated wastewater discharges. Environ. Pollut. 2018, 234, 538–542. [Google Scholar] [CrossRef]
- Dolejska, M.; Villa, L.; Poirel, L.; Nordmann, P.; Carattoli, A. Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance–nodulation–cell division/multidrug efflux pump. J. Antimicrob. Chemother. 2013, 68, 34–39. [Google Scholar] [CrossRef] [Green Version]
- Kubasova, T.; Cejkova, D.; Matiasovicova, J.; Sekelova, Z.; Polansky, O.; Medvecky, M.; Rychlik, I.; Juricova, H. Antibiotic resistance, core-genome and protein expression in IncHI1 plasmids in Salmonella Typhimurium. Genome Biol. Evol. 2016, 8, 1661–1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawley, T.D.; Gilmour, M.W.; Gunton, J.E.; Tracz, D.M.; Taylor, D.E. Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 2003, 185, 581–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sennati, S.; Di Pilato, V.; Riccobono, E.; Di Maggio, T.; Villagran, A.L.; Pallecchi, L.; Bartoloni, A.; Rossolini, G.M.; Giani, T. Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J. Antimicrob. Chemother. 2017, 72, 2127–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, Z.; Dong, H.; Wang, M.; Qin, S.; Chen, S.; Li, R. Emergence of tet(X4)-positive hypervirulent Klebsiella pneumoniae of food origin in China. LWT 2023, 173, 114280. [Google Scholar] [CrossRef]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Lei, T.; Zhang, J.; Jiang, F.; He, M.; Zeng, H.; Chen, M.; Wu, S.; Wang, J.; Ding, Y.; Wu, Q. First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 2019, 308, 108290. [Google Scholar] [CrossRef] [PubMed]
- Sumbana, J.J.; Santona, A.; Fiamma, M.; Taviani, E.; Deligios, M.; Zimba, T.; Lucas, G.; Sacarlal, J.; Rubino, S.; Paglietti, B. Extraintestinal pathogenic Escherichia coli ST405 isolate coharboring blaNDM-5 and blaCTXM-15: A new threat in Mozambique. Microb. Drug Resist. 2021, 27, 1633–1640. [Google Scholar] [CrossRef]
- Turchi, B.; Dec, M.; Bertelloni, F.; Winiarczyk, S.; Gnat, S.; Bresciani, F.; Viviani, F.; Cerri, D.; Fratini, F. Antibiotic susceptibility and virulence factors in Escherichia coli from sympatric wildlife of the Apuan Alps regional park (Tuscany, Italy). Microb. Drug Resist. 2019, 25, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Tang, B.; Lin, J.; Ed-Dra, A.; Lin, H.; Wu, J.; Dong, Y.; Yang, H.; Yue, M. Genome assessment of carbapenem-and colistin-resistant Escherichia coli from patients in a sentinel hospital in China. Cells 2022, 11, 3480. [Google Scholar] [CrossRef]
- Montealegre, M.C.; Rodríguez, A.T.; Roy, S.; Hossain, M.I.; Islam, M.A.; Lanza, V.F.; Julian, T.R. High genomic diversity and heterogenous origins of pathogenic and antibiotic-resistant Escherichia coli in household settings represent a challenge to reducing transmission in low-income settings. Msphere 2020, 5, e00704-19. [Google Scholar] [CrossRef] [Green Version]
- Yatsuyanagi, J.; Saito, S.; Miyajima, Y.; Amano, K.-I.; Enomoto, K. Characterization of Atypical Enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J. Clin. Microbiol. 2003, 41, 2033–2039. [Google Scholar] [CrossRef] [Green Version]

| IDs | Antibiotics | 812A1-131 | |
|---|---|---|---|
| MIC (μg/mL) | ARGs | ||
| 1 | Ampicillin | >128 R | blaTEM-1B, blaTEM-141, blaTEM-206, blaCTX-M-55, blaOXA-10, blaCTX-M-14 |
| 2 | Amoxicillin/clavulanic acid | >128/64 R | |
| 3 | Cefotaxime | >8 R | blaCTX-M-55, blaCTX-M-14 |
| 4 | Meropenem | 1 S | / |
| 5 | Amikacin | >64 R | rmtB |
| 6 | Gentamicin | >32 R | aac(3)-IV, rmtB |
| 7 | Colistin | 1 S | / |
| 8 | Ceftiofur | >32 R | |
| 9 | Ciprofloxacin | >8 R | qnrS1, parC, gyrA |
| 10 | Trimethoprim/sulfamethoxazole | >0.5/9.5 R | sul2 |
| 11 | Tetracycline | >64 R | tet(X4), tet(A) |
| 12 | Tigecycline | >16 R | tet(X4) |
| 13 | Florfenicol | 128 R | floR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Huang, Y.; Guan, C.; Li, J.; Yang, H.; Zhao, G.; Liu, C.; Ma, J.; Tang, B. Characterization of an Escherichia coli Isolate Coharboring the Virulence Gene astA and Tigecycline Resistance Gene tet(X4) from a Dead Piglet. Pathogens 2023, 12, 903. https://doi.org/10.3390/pathogens12070903
Wang J, Huang Y, Guan C, Li J, Yang H, Zhao G, Liu C, Ma J, Tang B. Characterization of an Escherichia coli Isolate Coharboring the Virulence Gene astA and Tigecycline Resistance Gene tet(X4) from a Dead Piglet. Pathogens. 2023; 12(7):903. https://doi.org/10.3390/pathogens12070903
Chicago/Turabian StyleWang, Jianmei, Yuting Huang, Chunjiu Guan, Jie Li, Hua Yang, Guoping Zhao, Canying Liu, Jiangang Ma, and Biao Tang. 2023. "Characterization of an Escherichia coli Isolate Coharboring the Virulence Gene astA and Tigecycline Resistance Gene tet(X4) from a Dead Piglet" Pathogens 12, no. 7: 903. https://doi.org/10.3390/pathogens12070903





