Abstract
Pseudomonas aeruginosa, an opportunistic pathogen causing infections in immunocompromised patients, usually shows pronounced antimicrobial resistance. In recent years, the frequency of carbapenemases in P. aeruginosa has decreased, which allows use of new beta-lactams/combinations in antimicrobial therapy. Therefore, the in vitro evaluation of these drugs in contemporary isolates is warranted. We evaluated the antimicrobial susceptibility and genomic aspects of 119 clinical P. aeruginosa isolates from 24 different hospitals in Brazil in 2021–2022. Identification was performed via MALDI-TOF-MS, and antimicrobial susceptibility was identified through broth microdilution, gradient tests, or disk diffusion. Whole-genome sequencing was carried out using NextSeq equipment. The most active drug was cefiderocol (100%), followed by ceftazidime–avibactam (94.1%), ceftolozane–tazobactam (92.4%), and imipenem–relebactam (81.5%). Imipenem susceptibility was detected in 59 isolates (49.6%), and the most active aminoglycoside was tobramycin, to which 99 (83.2%) isolates were susceptible. Seventy-one different sequence types (STs) were detected, including twelve new STs described herein. The acquired resistance genes blaCTX-M-2 and blaKPC-2 were identified in ten (8.4%) and two (1.7%) isolates, respectively. Several virulence genes (exoSTUY, toxA, aprA, lasA/B, plcH) were also identified. We found that new antimicrobials are effective against the diverse P. aeruginosa population that has been circulating in Brazilian hospitals in recent years.
Keywords:
CAZ-AVI; MEM-VAR; cefiderocol; fosfomycin; polymyxin; whole genome sequencing; Illumina; MLST 1. Introduction
Pseudomonas aeruginosa ranks as the most frequent non-fermentative Gram-negative bacterium associated with hospital-acquired infections, mainly affecting seriously ill patients in intensive care units (ICUs) [1]. It is part of the ESKAPE group, which comprises drug-resistant pathogens of clinical importance, namely Enterococcus, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter [2]. P. aeruginosa presenting carbapenem resistance is listed as one of the critical pathogens defined by the World Health Organization and as a serious threat according to the Centers for Disease Control and Prevention [3,4]. In the United States, the rate of hospital-onset multidrug-resistant (MDR) P. aeruginosa infections increased by more than 30% in 2020 compared to 2019, mainly due to the COVID-19 pandemic [5]. In Brazil, high endemic rates of drug-resistant pathogens are reported [6,7]. Regarding P. aeruginosa specifically, the SPM-1-producing, colistin-only susceptible (COS) ST277 clone has been reported to predominate in the country throughout the last two decades [8]. Over the years, surveillance studies have shown stability in the rates of carbapenem-resistance in P. aeruginosa from Latin America [9]. Nevertheless, the frequency of SPM-producing P. aeruginosa has been consistently reported to be decreasing in recent years [10,11,12], showing the involvement of non-enzymatic mechanisms in carbapenem resistance, including porin loss and efflux pump system overproduction [13]. This change in the epidemiology of carbapenem resistance allows for the opportunity to use new beta-lactams/combinations in the treatment of P. aeruginosa infections. Therefore, it is necessary to perform an in vitro evaluation of these drugs using contemporary isolates from clinical sources. In this study, we aimed to evaluate a large collection of clinical P. aeruginosa isolates recovered from various hospitals in Brazil during 2021–2022. The whole-genome sequence of the isolates was obtained in order to define their clonality, resistome, and virulome characteristics.
2. Materials and Methods
2.1. Isolate Selection and Identification
On a continuous and voluntary basis, the Instituto Adolfo Lutz receives clinical isolates of hospitalized patients presenting infections, associated with outbreaks or not, for phenotypic and genotypic antimicrobial resistance characterization. Bacterial identification was initially carried out via phenotypic testing and MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). Between January 2021 and August 2022, 216 isolates identified as P. aeruginosa were received in our laboratory. A total of 70 isolates recovered from non-human sources (hospital environment) or redundant isolates from the same patient (recovered within one month) were excluded; thus, 146 isolates were analyzed.
2.2. Antimicrobial Susceptibility Testing
The isolates were initially processed using disk-diffusion antimicrobial susceptibility testing for amikacin, aztreonam, cefepime, ceftazidime, ciprofloxacin, doripenem, gentamicin, imipenem, levofloxacin, meropenem, netilmicin, piperacillin-tazobactam, ticarcillin-clavulanate, and tobramycin with Oxoid (Basingstoke, United Kingdom) disks. Complementarily, an in-house broth microdilution method using cation-adjusted Mueller–Hinton Broth (Sigma-Aldrich, St. Louis, MO, USA) was performed to determine the minimum inhibitory concentration (MIC) for amikacin gentamicin, imipenem, meropenem, colistin, polymyxin B, tigecycline, and ceftazidime-avibactam, and all salts were purchased from Sigma-Aldrich (St. Louis, MO, USA), except for avibactam, which was donated by Pfizer Inc. To complete the antimicrobial susceptibility panel, novel antimicrobials/combinations were evaluated with Liofilchem (Roseto degli Abruzzi, Italy) MIC test strips for ceftolozane–tazobactam, meropenem–vaborbactam, imipenem–relebactam, cefoperazone–sulbactam, cefiderocol, plazomicin, eravacycline, and fosfomycin.
2.3. Phenotypic and Genotypic Carbapenemase Detection
We used a Fourier test to detect the production of carbapenemases [14]. This test consists of applying different amounts of cloxacillin salt in imipenem disks to discriminate between carbapenemase-producing and non-producing strains of P. aeruginosa. Isolates presenting differences in imipenem halos with and without cloxacillin ≤ 5 mm were identified as carbapenemase producers. We excluded isolates producing metallo-β-lactamases (MBL) detected via multiplex PCR targeting the main genes blaNDM, blaSPM, blaIMP, and blaVIM [15].
2.4. DNA Extraction, Whole Genome Sequencing, and Assembly
Whole bacterial DNA was extracted by using Invitrogen PureLink Genomic DNA Mini Kit (USA) following the manufacturer’s recommendations. After extraction, the DNA was quantified using a Qubit (Thermo Scientific Inc., Waltham, MA, USA) fluorometer and the libraries were prepared for Illumina (USA) NextSeq sequencing, using a P1/300 cycle cartridge. The library preparation and Illumina runs were performed at the Strategic Laboratory, Instituto Adolfo Lutz, São Paulo, Brazil. After the evaluation of quality control parameters (read sizes, Phred values > 30, GC content), genomes were de novo assembled using CLC Workbench software (Qiagen, Germany).
2.5. Annotation, Resistome and Virulome Detection, Serotype Prediction, and MLST
The assembled genomes were uploaded to the Galaxy Europe platform [16] and then annotated with Prokka [17]. Acquired resistance and virulence-codifying genes were detected with Abricate using the Resfinder [18] and VirulenceFinder [19] databases, respectively. Chromosomal mutations leading to antimicrobial resistance were detected using Pointfinder software [20]. The in silico serotype was determined via the pseudomonas aeruginosa serotyper (PAst) program [21] available at the Center for Genomic Epidemiology webserver (https://www.genomicepidemiology.org/, accessed on 22 April 2023). Sequence types (STs) were defined based on the internal sequences of seven housekeeping genes [22]. When new alleles or allele combinations were identified, the isolates were submitted to pubMLST for curation and assignment [23].
2.6. Phylogenetic Analysis
The Prokka-annotated genomes were used to generate a core genome alignment with Roary v3.13.0 [24]. Core genome phylogeny was inferred from the core genome alignment, and a maximum-likelihood tree was constructed using IQ-TREE [25] v.2.1.2 with standard model selection followed by tree inference and 1000 bootstrap replicates. The tree was visualized in the Microreact platform [26] and can be interactively accessed at https://microreact.org/project/ogubUpsSXzEseq311xMeth-pseudomonas-non-mbl-119 (accessed on 22 April 2023).
3. Results
Initially, 146 isolates were enrolled in this study (75 of them were imipenem resistant), but 12 were excluded because they were MBL-producers (MBL frequency among the imipenem-resistant isolates was 16%), namely 6 blaSPM, 4 blaVIM, and 2 blaIMP isolates. In addition, quality parameters were not achieved after the sequencing of 15 isolates, which were excluded. Therefore, 119 isolates recovered from the clinical specimens of 115 non-repetitive patients who attended one of twenty-four public or private health institutions located in 16 different Brazilian cities, were included. The isolates were recovered mainly from the respiratory tract (n = 58; 48.7%), followed by blood or central venous catheter tips (26; 21.8%), urine (16; 13.4%), infected wounds (6; 5.0%), surveillance swabs (3; 2.5%), or cerebrospinal fluid (1, 0.8%), and nine isolates (7.6%) were recovered from other clinical sources.
Antimicrobial susceptibility evaluated via disk-diffusion is presented in Table 1. According to the Magiorakus classification, most of the isolates were identified as multidrug-resistant (MDR) (55.5%) or extensively drug-resistant (XDR) (18.5%), and one isolate (0.8%) was identified as pan-drug-resistant (PDR). The remaining 30 isolates (25.2%) were classified as susceptible.

Table 1.
Antimicrobial susceptibility of clinical isolates of P. aeruginosa from Brazil recovered between 2021 and 2022 (n = 119).
MIC values determined via broth-microdilution or gradient tests (Table 2) showed that all isolates were susceptible to cefiderocol (100% susceptibility), and the most active drugs were ceftazidime–avibactam (94.1%), ceftolozane–tazobactam (92.4%), and imipenem–relebactam (81.5%). Among classical drugs, a comparable susceptibility rate was found only for tobramycin (83.2%). The carbapenem susceptibility rates ranged from 47.9% (imipenem, broth microdilution) to 66.4% (doripenem, disk-diffusion). Resistance to colistin and polymyxin B was identified in four (3.4%) and three (2.5%) isolates, respectively; according to CLSI, the remaining isolates were classified as intermediate (I). The distribution of MIC values, as well as the MIC50/MIC90 values, demonstrated the inefficacy of tigecycline, fosfomycin, and eravacycline against these P. aeruginosa isolates (Table 2).

Table 2.
Antimicrobial susceptibility (quantitative results) of Pseudomonas aeruginosa isolates from Brazil (n = 119). The table indicates the breakpoints for susceptibility categorization and the reference from which those breakpoints were retrieved. The minimal inhibitory concentration (MIC) values (μg/mL) for inhibiting the growth of 50% (MIC50, bold) and 90% (MIC90, underlined) of the isolates are indicated, as is the cumulative distribution of isolates.
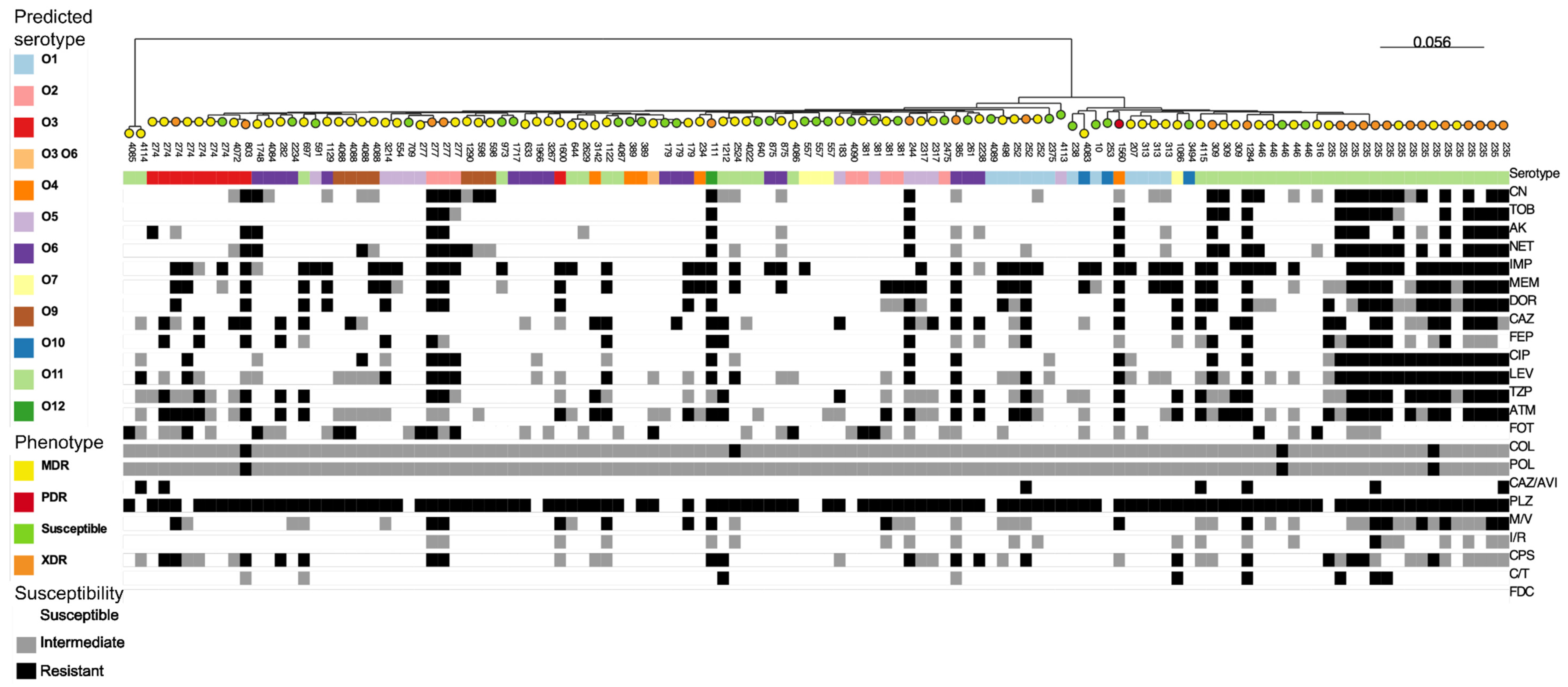
Genomic analysis identified 71 different sequence types (STs) among the 119 P. aeruginosa sequences evaluated. Of note, twelve new STs were identified for the first time in this study, of which eight are new allele combinations and three are new alleles (mutL, aroE, trpE)—one isolate presented new alleles for two genes simultaneously (guaA and mutL). The most frequent ST, ST235, was found in sixteen isolates (13.4%) from eight hospitals across seven cities; this ST was identified in isolates recovered from different sources, including blood, infected wounds, respiratory tracts, and surveillance swabs. The distribution of common STs (detected in more than one isolate), according to hospital and isolation source, is presented in Table 3. An in silico analysis identified eleven predicted serotypes, with O11 (33.6%) and O1 (10.1%) as the most frequent, and all the ST235 isolates (n = 16) presented the O11 predicted serotype (Table S1). The phylogenetic tree based on pangenome analysis correlated with STs and partially with the predicted serotypes, showing that the ST235 isolates to be more resistant than isolates with other STs (Figure 1; Table S2). In fact, the XDR phenotype was more frequent in ST235 isolates (12/16, 75%) than in isolates belonging to other STs (10/103, 10.7%) (p < 0.0001 on the Yates’ chi-squared test).

Table 3.
Distribution of common ST (>1 isolate) clinical isolates of P. aeruginosa from Brazil recovered between 2021 and 2022 (n = 119), according to municipalities, hospitals, and source of isolation.

Figure 1.
Maximum-likelihood tree generated through core genome alignment with Roary followed by IQ-TREE analysis in 119 isolates (Table S2) of clinical P. aeruginosa from Brazil. In this tree, the ST (indicated by the number for each strain), predicted serotype (colored squares), antimicrobial resistance (white, gray, or black squares), and resistance phenotypes (colored circles) are represented. The tree can be interactively accessed on the link https://microreact.org/project/ogubUpsSXzEseq311xMeth-pseudomonas-non-mbl-119 (accessed on 22 April 2023).
In addition to the chromosomally and naturally encoded β-lactamases ampC and blaOXA, acquired antimicrobial resistance genes conferring resistance to beta-lactams were identified. Nine isolates (eight with the XDR phenotype and one MDR) carried the ESBL blaCTX-M-2 gene, and they were identified as ST235 (n = 6), ST111, ST244, and ST309 (n = 1, each). The blaGES-1 gene was identified in another ST235 XDR isolate, and the blaKPC-2 gene was found in one XDR isolate (ID_0367_21) belonging to ST803. One isolate (ID_0455_22), XDR, ST1284, simultaneously carried the tetG, blaKPC-2 and blaCTX-M-2 genes. The analysis of the genetic environment associated with the blaKPC-2 gene showed that isolate ID_0367_21 carries the gene on a plasmid with a DNA sequence similar to plasmids harboring carbapenemases that circulate in P. aeruginosa worldwide (corresponding to GenBank Accession Numbers CP078000, LC586269, CP077989). On this plasmid, the gene is located in a truncated non-Tn4401 genetic element (NTEKPC), similar to that presented in FII-FIB(pQil) plasmids that harbor KPC-3 in Klebsiella pneumoniae isolates from the USA [27]. In contrast, it was not possible to define whether the blaKPC-2 gene of isolate ID_0455_22 is present on a plasmid or the chromosome. However, blaKPC-2 is present in a truncated form of Tn4401 element isoform b. Our analysis revealed that a segment of 6844 bp with 99.9% similarity to Tn4401b and an intact IRR is present in the genome of this isolate. Another MDR isolated with ST309 was found to carry the tetG gene. Several genes that confer resistance to aminoglycosides were identified, such as aac, aad, ant, and aph; conversely, armA or rmt-family genes, which are associated with high levels of aminoglycoside resistance, were not detected.
Genes associated with the four effector proteins of the type III secretion system (T3SS), namely exoS, exoT, exoU, and exoY, were detected at frequencies of 63.8%, 98.3%, 30.2%, and 92.4%, respectively. As expected [28], the association of exoS and exoU in the same isolate was not detected. Additionally, genes encoding exotoxin A (toxA) were detected in 97.5% of the isolates; alkaline protease (aprA), elastase (lasA/B), and phospholipase C (plcH) were detected in all of the isolates (except for one isolate negative for the lasA gene).
4. Discussion
Although studies have focused on changes in P. aeruginosa antimicrobial susceptibility, only scarce information is available for genomics findings for this species, particularly for specimens with the MBL-negative phenotype, which is increasing in Brazilian settings.
In this study, we identified that antimicrobial agents not in use in clinical settings have preserved activity against clonally diverse P. aeruginosa clinical isolates. P. aeruginosa is recognized as an opportunistic pathogen causing skin and soft tissue infections and even potentially fulminant invasive infections [29]. The treatment of P. aeruginosa infections is limited because of both intrinsic and acquired resistance mechanisms [30]. Carbapenems have been employed for clinical treatment, but with the emergence of metallo-beta-lactamases, a potent class of carbapenemases, beta-lactams (with the exception of aztreonam, which is not hydrolyzed by MBL) are no longer recommended for the treatment of P. aeruginosa [30]. It is known that difficult-to-treat infections due to P. aeruginosa result in worse prognosis with poor outcomes [1].
Molecular studies have identified that the spread of MBL production in Brazil is associated with a dissemination of a specific sequence type, ST277, the so-called colistin-only susceptible (COS) P. aeruginosa [31]. More recently, however, a reduction in the prevalence of MBL-producing ST277 was observed in Brazilian hospitals, but the reasons for this change in epidemiology are not clear [10,11,12]. By analyzing a large collection of P. aeruginosa recovered from the clinical specimens of patients admitted to different hospitals, we identified 71 different STs among the 119 isolates evaluated. The diversity of clones, which has been reported in previous smaller studies in recent years, may be linked to the widespread misuse of antimicrobials rather than clonal dissemination through cross-transmission [32].
Intriguingly, we identified that ST235 isolates carried more resistance markers than isolates with other STs. ST235 is considered a “high-risk clone” because of its ability to accumulate antimicrobial resistance genes and is widespread in diverse hospital settings [33]. We speculate that a transition from the ST277 to the ST235 P. aeruginosa clone, which is still expanding in Brazilian hospitals, has been occurring. As ST235 was found to display more resistance than the other clones (Figure 1), attention must be paid to this high-risk clone to avoid or at least reduce its dissemination.
We identified the high activity of cefiderocol (100%), ceftazidime–avibactam (94.1%), ceftolozane–tazobactam (92.4%), and imipenem–relebactam (81.5%) in our analyses. Of these drugs, only cefiderocol is currently not approved for the treatment of P. aeruginosa infections in Brazil. Avibactam and relebactam are second-generation beta-lactamase inhibitors able to inhibit class A, C, and D beta-lactamases, with activity against KPC-producing Enterobacterales and KPC-producing Pseudomonas [34]. In our study, two isolates with KPC-2 in different genetic backgrounds were identified, which is not common in our region [35] but has been reported in other countries [36,37,38,39].
Regarding currently available drugs in hospital settings, polymyxin B and colistin were found to present high activity against the P. aeruginosa isolates, as observed by the MIC50 and MIC90 results (Table 2). In a realistic scenario, empiric treatment with polymyxins is still employed in Brazilian settings, which, in theory, would cover the contemporary P. aeruginosa isolates causing hospital-acquired infections in the country. Nevertheless, recent guidelines do not recommend use of this class of drugs for severe infections caused by carbapenem-resistant P. aeruginosa, as some studies have shown worse outcomes, especially when compared to ceftolozane–tazobactam [40]. Molecular analysis identified that only 16% of the carbapenem resistance in P. aeruginosa is mediated by metallo-beta-lactamase production, rendering the use of ceftolozane–tazobactam, ceftazidime–avibactam, or other beta-lactam combinations instead of polymyxins possible as empirical treatments in Brazilian hospitals.
In addition to the robust data on antimicrobial susceptibility generated in this study, the determination of the resistome and virulome for each isolate is notable. The inclusion of such genomes in public databases will contribute to future studies with both local and global perspectives, allowing for the tracking of an important public health pathogen of clinical relevance.
5. Conclusions
In summary, by analyzing 119 P. aeruginosa isolates with diverse genetic backgrounds from 24 Brazilian hospitals, we identified the preserved activity of antimicrobial agents with restricted its use in our country. The continuous investigation of antimicrobial susceptibility, associated with the rational use of antimicrobial agents in clinical practice, is essential to preserving the scarce options for the treatment of bacterial pathogens causing health care-associated infections.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pathogens12070918/s1, Table S1: Heatmap showing the distribution of predicted serotypes according to ST; Table S2: List of strains sequenced in this study and additional metadata.
Author Contributions
Conceptualization, C.H.C. and P.B.; methodology, C.H.C., A.Y.Y., A.R.d.S., M.d.J.d.C.L., P.S.P.F., M.B.N.d.S. and K.R.C.; software, C.H.C., M.R.T.-C. and M.P.V.C.; formal analysis, C.H.C., C.T.S., M.P.F. and P.B.; resources, C.H.C. and P.B.; data curation, C.H.C. and C.T.S.; writing—original draft preparation, C.H.C., M.P.V.C. and M.P.F.; writing—review and editing, M.R.T.-C. and P.B.; project administration and funding acquisition, P.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an Investigator-Sponsored Research grant from Pfizer/Wyeth. Authors are thankful to FAPESP for indirectly supporting this study via the grants 2023/08958-0, 2019/21361-8, 2017/50333-7, and 2018/21192-9 from the São Paulo Research Foundation (Fundação de Amparo à Pesquisa do Estado de São Paulo). CHC received a Productivity Research Fellowship and grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 302543/2021-0 and CNPq 402158/2021-0).
Institutional Review Board Statement
This study was submitted to and approved by the local ethics committee (CAAE 56976022.2.0000.0059) and scientific board (CTC-27N-2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequences generated in this study were deposited in pubMLST and NCBI/Genbank platforms (Table S2).
Acknowledgments
This work was supported by an Investigator-Sponsored Research grant from Pfizer/Wyeth. The authors are thankful to FAPESP for indirectly supporting this study via the grants 2019/21361-8, 2017/50333-7, 2018/21192-9, São Paulo Research Foundation (Fundação de Amparo à Pesquisa do Estado de São Paulo). CHC received a Productivity Research Fellowship and grants from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 302543/2021-0 and CNPq 402158/2021-0). The authors also acknowledge the financial support of FESIMA (Fundo Especial de Saúde para Imunização em Massa e Controle de Doenças) for AYY and ARS. The funding agencies played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. This study was submitted to and approved by the local ethics committee (CAAE 56976022.2.0000.0059) and scientific board CTC-27N-2021).
Conflicts of Interest
PB received an Investigator-Sponsored Research grant from Pfizer/Wyeth. CHC and MRT have received Productivity Research Fellowships from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). The remaining authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-Resistant Pseudomonas aeruginosa: Clinical Impact and Complex Regulation of Chromosomally Encoded Resistance Mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical Relevance of the ESKAPE Pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2013; pp. 1–113. [CrossRef]
- Centers for Disease Control and Prevention. 2022 Special Report: COVID-19 U.S. Impact on Antimicrobial Resistance. Available online: https://stacks.cdc.gov/view/cdc/117915 (accessed on 8 March 2023).
- Rossi, F. The Challenges of Antimicrobial Resistance in Brazil. Clin. Infect. Dis. 2011, 52, 1138–1143. [Google Scholar] [CrossRef] [PubMed]
- Fortaleza, C.M.C.B.; Padoveze, M.C.; Kiffer, C.R.V.; Barth, A.L.; Carneiro, I.C.D.R.S.; Giamberardino, H.I.G.; Rodrigues, J.L.N.; Santos Filho, L.; de Mello, M.J.G.; Pereira, M.S.; et al. Multi-State Survey of Healthcare-Associated Infections in Acute Care Hospitals in Brazil. J. Hosp. Infect. 2017, 96, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.M.; Carmo, M.S.; Silbert, S.; Gales, A.C. SPM-1-Producing Pseudomonas aeruginosa: Analysis of the Ancestor Relationship Using Multilocus Sequence Typing, Pulsed-Field Gel Electrophoresis, and Automated Ribotyping. Microb. Drug Resist. 2011, 17, 215–220. [Google Scholar] [CrossRef]
- Shortridge, D.; Gales, A.C.; Streit, J.M.; Huband, M.D.; Tsakris, A.; Jones, R.N. Geographic and Temporal Patterns of Antimicrobial Resistance in Pseudomonas aeruginosa over 20 Years from the SENTRY Antimicrobial Surveillance Program, 1997–2016. Open Forum Infect. Dis. 2019, 6, S63–S68. [Google Scholar] [CrossRef]
- Campana, E.H.; Xavier, D.E.; Petrolini, F.V.-B.B.; Cordeiro-Moura, J.R.; de Araujo, M.R.E.; Gales, A.C. Carbapenem-Resistant and Cephalosporin-Susceptible: A Worrisome Phenotype among Pseudomonas aeruginosa Clinical Isolates in Brazil. Braz. J. Infect. Dis. 2017, 21, 57–62. [Google Scholar] [CrossRef]
- Cacci, L.C.; Chuster, S.G.; Martins, N.; do Carmo, P.R.; Girão, V.B.d.C.; Nouér, S.A.; de Freitas, W.V.; de Matos, J.A.; Magalhães, A.C.d.G.; Ferreira, A.L.P.; et al. Mechanisms of Carbapenem Resistance in Endemic Pseudomonas aeruginosa Isolates after an SPM-1 Metallo-β-Lactamase Producing Strain Subsided in an Intensive Care Unit of a Teaching Hospital in Brazil. Mem. Inst. Oswaldo Cruz 2016, 111, 551–558. [Google Scholar] [CrossRef]
- de Sá Cavalcanti, F.L.; Almeida, A.C.S.; Vilela, M.A.; de Morais, M.M.C.; de Morais, M.A. Changing the Epidemiology of Carbapenem-Resistant Pseudomonas aeruginosa in a Brazilian Teaching Hospital: The Replacement of São Paulo Metallo-β-Lactamase-Producing Isolates. Mem. Inst. Oswaldo Cruz 2012, 107, 420–423. [Google Scholar] [CrossRef]
- Castanheira, M.; Mills, J.C.; Farrell, D.J.; Jones, R.N. Mutation-Driven β-Lactam Resistance Mechanisms among Contemporary Ceftazidime-Nonsusceptible Pseudomonas aeruginosa Isolates from U.S. Hospitals. Antimicrob. Agents Chemother. 2014, 58, 6844–6850. [Google Scholar] [CrossRef]
- Fournier, D.; Garnier, P.; Jeannot, K.; Mille, A.; Gomez, A.-S.; Plésiat, P. A Convenient Method To Screen for Carbapenemase-Producing Pseudomonas aeruginosa. J. Clin. Microbiol. 2013, 51, 3846. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for Detection of Acquired Carbapenemase Genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Nekrutenko, A.; Grüning, B.A.; Blankenberg, D.; Goecks, J.; Schatz, M.C.; Ostrovsky, A.E.; Mahmoud, A.; Lonie, A.J.; Syme, A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2022 Update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A Novel Web Tool for WGS-Based Detection of Antimicrobial Resistance Associated with Chromosomal Point Mutations in Bacterial Pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Thrane, S.W.; Taylor, V.L.; Lund, O.; Lam, J.S.; Jelsbak, L. Application of Whole-Genome Sequencing Data for O-Specific Antigen Analysis and In Silico Serotyping of Pseudomonas aeruginosa Isolates. J. Clin. Microbiol. 2016, 54, 1782–1788. [Google Scholar] [CrossRef]
- Curran, B.; Jonas, D.; Grundmann, H.; Pitt, T.; Dowson, C.G. Development of a Multilocus Sequence Typing Scheme for the Opportunistic Pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004, 42, 5644–5649. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications [Version 1; Referees: 2 Approved]. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Argimón, S.; Abudahab, K.; Goater, R.J.E.; Fedosejev, A.; Bhai, J.; Glasner, C.; Feil, E.J.; Holden, M.T.G.; Yeats, C.A.; Grundmann, H.; et al. Microreact: Visualizing and Sharing Data for Genomic Epidemiology and Phylogeography. Microb. Genom. 2016, 2, e000093. [Google Scholar] [CrossRef] [PubMed]
- Shropshire, W.C.; Dinh, A.Q.; Earley, M.; Komarow, L.; Panesso, D.; Rydell, K.; Gómez-Villegas, S.I.; Miao, H.; Hill, C.; Chen, L.; et al. Accessory Genomes Drive Independent Spread of Carbapenem- Resistant Klebsiella pneumoniae Clonal Groups 258 and 307 in Houston, TX. MBio 2022, 13, e00497-22. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.R. The Type III Secretion System of Pseudomonas aeruginosa: Infection by Injection. Nat. Rev. Microbiol. 2009, 7, 654. [Google Scholar] [CrossRef] [PubMed]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas Genomes: Diverse and Adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Fonseca, E.L.; Freitas, F.D.; Vicente, A.C. The Colistin-Only-Sensitive (COS) Brazilian Pseudomonas aeruginosa Clone SP (ST 277) Is Spread Worldwide. Antimicrob. Agents Chemother. 2010, 54, 2743. [Google Scholar] [CrossRef]
- El-Basst, R.; Saliba, S.; Saleh, L.; Saoud, N.; Azar, E.; Zalloua, P.; Chamieh, A. The Effect of Decreased Antipseudomonal Drug Consumption on Pseudomonas aeruginosa Incidence and Antimicrobial Susceptibility Profiles over 9 Years in a Lebanese Tertiary Care Center. Antibiotics 2023, 12, 192. [Google Scholar] [CrossRef]
- Oliver, A.; Mulet, X.; López-Causapé, C.; Juan, C. The Increasing Threat of Pseudomonas aeruginosa High-Risk Clones. Drug Resist. Updat. 2015, 21–22, 41–59. [Google Scholar] [CrossRef]
- Zasowski, E.J.; Rybak, J.M.; Rybak, M.J. The β-Lactams Strike Back:Ceftazidime-Avibactam. Pharmacotherapy 2015, 35, 755. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.L.d.C.; Ximenes, R.M.; Maciel, M.A.V. Occurrence of BlaKPC Gene in Clinical Isolates of Pseudomonas aeruginosa from Brazil. ABCS Health Sci. 2022, 47, e022306. [Google Scholar] [CrossRef]
- Santella, G.; Cittadini, R.; Papalia, M.; Vera Ocampo, C.; Del Castillo, M.; Vay, C.; Gutkind, G.; Radice, M. First Clonal Spread of KPC-Producing Pseudomonas aeruginosa in Buenos Aires, Argentina. Infect. Genet. Evol. 2012, 12, 2003–2005. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, C.; Wang, Q.; Zeng, Y.; Sun, Q.; Shu, L.; Lu, J.; Cai, J.; Wang, S.; Zhang, R.; et al. Emergence and Expansion of a Carbapenem-Resistant Pseudomonas aeruginosa Clone Are Associated with Plasmid-Borne Bla KPC-2 and Virulence-Related Genes. mSystems 2021, 6, e00154-21. [Google Scholar] [CrossRef]
- Carrara-Marroni, F.E.; Cayô, R.; Streling, A.P.; Da Silva, A.C.R.; Palermo, R.L.; Romanin, P.; Venâncio, E.; Perugini, M.R.E.; Pelisson, M.; Gales, A.C. Emergence and Spread of KPC-2-Producing Pseudomonas aeruginosa Isolates in a Brazilian Teaching Hospital. J. Glob. Antimicrob. Resist. 2015, 3, 304–306. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-Producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 2010, 54, 3072. [Google Scholar] [CrossRef]
- Paul, M.; Carrara, E.; Retamar, P.; Tängdén, T.; Bitterman, R.; Bonomo, R.A.; de Waele, J.; Daikos, G.L.; Akova, M.; Harbarth, S.; et al. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Guidelines for the Treatment of Infections Caused by Multidrug-Resistant Gram-Negative Bacilli (Endorsed by European Society of Intensive Care Medicine). Clin. Microbiol. Infect. 2022, 28, 521–547. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).