Genomic Analysis of Yersinia pestis Strains from Brazil: Search for Virulence Factors and Association with Epidemiological Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Data
2.2. Evaluation of Genome Sequencing
2.3. Genome Assembly and Annotation
2.4. Pangenome Analysis
2.5. Statistical Analysis of Presence and Absence Genes in Pangenomics
2.6. Identification of Virulence Factors
2.7. Association of Pangenome Data and Epidemiological Data
2.8. Core Genome (cg) Multilocus Sequence Typing (MLST)
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbieri, R.; Signoli, M.; Chevé, D.; Costedoat, C.; Tzortzis, S.; Aboudharam, G.; Raoult, D.; Drancourt, M. Yersinia Pestis: The Natural History of Plague. Clin. Microbiol. Rev. 2020, 34, e00044-19. [Google Scholar] [CrossRef] [PubMed]
- Bramanti, B.; Stenseth, N.C.; Walløe, L.; Lei, X. Plague: A Disease Which Changed the Path of Human Civilization. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2016; Volume 918, pp. 1–26. [Google Scholar] [CrossRef]
- Vogler, A.J.; Sahl, J.W.; Leal, N.C.; Sobreira, M.; Williamson, C.H.D.; Bollig, M.C.; Birdsell, D.N.; Rivera, A.; Thompson, B.; Nottingham, R.; et al. A Single Introduction of Yersinia Pestis to Brazil during the 3rd Plague Pandemic. PLoS ONE 2019, 14, e0209478. [Google Scholar] [CrossRef] [PubMed]
- da Silva Fernandes, D.L.R.; de Souza Gomes, E.C.; Bezerra, M.F.; de Paula Souza E Guimarães, R.J.; de Almeida, A.M.P. Spatiotemporal Analysis of Bubonic Plague in Pernambuco, Northeast of Brazil: Case Study in the Municipality of Exu. PLoS ONE 2021, 16, e0249464. [Google Scholar] [CrossRef]
- Giles, J.; Peterson, A.T.; Almeida, A. Ecology and Geography of Plague Transmission Areas in Northeastern Brazil. PLoS Negl. Trop. Dis. 2011, 5, e925. [Google Scholar] [CrossRef] [PubMed]
- Vallès, X.; Stenseth, N.C.; Demeure, C.; Horby, P.; Mead, P.S.; Cabanillas, O.; Ratsitorahina, M.; Rajerison, M.; Andrianaivoarimanana, V.; Ramasindrazana, B.; et al. Human Plague: An Old Scourge That Needs New Answers. PLoS Negl. Trop. Dis. 2020, 14, e0008251. [Google Scholar] [CrossRef]
- Bertherat, E. Plague around the World in 2019. Wkly. Epidemiol. Rec. = Relevé Épidémiologique Hebd. 2019, 94, 289–292. [Google Scholar]
- Perry, R.D.; Fetherston, J.D. Yersinia Pestis—Etiologic Agent of Plague. Clin. Microbiol. Rev. 1997, 10, 35–66. [Google Scholar] [CrossRef]
- Deng, W.; Burland, V.; Plunkett, G.; Boutin, A.; Mayhew, G.F.; Liss, P.; Perna, N.T.; Rose, D.J.; Mau, B.; Zhou, S.; et al. Genome Sequence of Yersinia Pestis KIM. J. Bacteriol. 2002, 184, 4601–4611. [Google Scholar] [CrossRef]
- Parkhill, J.; Wren, B.W.; Thomson, N.R.; Titball, R.W.; Holden, M.T.G.; Prentice, M.B.; Sebaihia, M.; James, K.D.; Churcher, C.; Mungall, K.L.; et al. Genome Sequence of Yersinia Pestis, the Causative Agent of Plague. Nature 2001, 413, 523–527. [Google Scholar] [CrossRef]
- Keller, M.; Spyrou, M.A.; Scheib, C.L.; Neumann, G.U.; Kröpelin, A.; Haas-Gebhard, B.; Päffgen, B.; Haberstroh, J.; Ribera i Lacomba, A.; Raynaud, C.; et al. Ancient Yersinia Pestis Genomes from across Western Europe Reveal Early Diversification during the First Pandemic (541–750). Proc. Natl. Acad. Sci. USA 2019, 116, 12363–12372. [Google Scholar] [CrossRef]
- Rascovan, N.; Sjögren, K.-G.; Kristiansen, K.; Nielsen, R.; Willerslev, E.; Desnues, C.; Rasmussen, S. Emergence and Spread of Basal Lineages of Yersinia Pestis during the Neolithic Decline. Cell 2019, 176, 295–305.e10. [Google Scholar] [CrossRef]
- Andrades Valtueña, A.; Neumann, G.U.; Spyrou, M.A.; Musralina, L.; Aron, F.; Beisenov, A.; Belinskiy, A.B.; Bos, K.I.; Buzhilova, A.; Conrad, M.; et al. Stone Age Yersinia Pestis Genomes Shed Light on the Early Evolution, Diversity, and Ecology of Plague. Proc. Natl. Acad. Sci. USA 2022, 119, e2116722119. [Google Scholar] [CrossRef]
- Bland, D.M.; Miarinjara, A.; Bosio, C.F.; Calarco, J.; Hinnebusch, B.J. Acquisition of Yersinia Murine Toxin Enabled Yersinia Pestis to Expand the Range of Mammalian Hosts That Sustain Flea-Borne Plague. PLoS Pathog. 2021, 17, e1009995. [Google Scholar] [CrossRef]
- Hinnebusch, B.J.; Rudolph, A.E.; Cherepanov, P.; Dixon, J.E.; Schwan, T.G.; Forsberg, Å. Role of Yersinia Murine Toxin in Survival of Yersinia Pestis in the Midgut of the Flea vector. Science 2002, 296, 733–735. [Google Scholar] [CrossRef]
- Leal, N.C.; Sobreira, M.; Araújo, A.F.Q.; Magalhães, J.L.O.; Vogler, A.J.; Bollig, M.C.; Nottingham, R.; Keim, P.; Wagner, D.M.; Almeida, A.M.P. Viability of Yersinia Pestis Subcultures in Agar Stabs. Lett. Appl. Microbiol. 2016, 62, 91–95. [Google Scholar] [CrossRef]
- Rocha, A.; Barbosa, C.S.; Brandão Filho, S.P.; Oliveira, C.M.F.d.; Almeida, A.M.P.d.; Gomes, Y.d.M. Primeiro Workshop Interno Dos Serviços de Referência Do Centro de Pesquisas Aggeu Magalhães Da Fundação Oswaldo Cruz. Rev. Patol. Trop./J. Trop. Pathol. 2010, 38, 299–309. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 April 2020).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de Novo Short Read Assembly Using de Bruijn Graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Gremme, G.; Steinbiss, S.; Kurtz, S. GenomeTools: A Comprehensive Software Library for Efficient Processing of Structured Genome Annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 645–656. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating Signal Peptides from Transmembrane Regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Brynildsrud, O.; Bohlin, J.; Scheffer, L.; Eldholm, V. Rapid Scoring of Genes in Microbial Pan-Genome-Wide Association Studies with Scoary. Genome Biol. 2016, 17, 238. [Google Scholar] [CrossRef]
- Chen, L. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2004, 33, D325–D328. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST plus: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. ChewBBACA: A Complete Suite for Gene-by-Gene Schema Creation and Strain Identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Zhou, D.; Han, Y.; Song, Y.; Tong, Z.; Wang, J.; Guo, Z.; Pei, D.; Pang, X.; Zhai, J.; Li, M.; et al. DNA Microarray Analysis of Genome Dynamics in Yersinia Pestis: Insights into Bacterial Genome Microevolution and Niche Adaptation. J. Bacteriol. 2004, 186, 5138–5146. [Google Scholar] [CrossRef]
- Demeure, C.E.; Dussurget, O.; Mas Fiol, G.; Le Guern, A.-S.; Savin, C.; Pizarro-Cerdá, J. Yersinia Pestis and Plague: An Updated View on Evolution, Virulence Determinants, Immune Subversion, Vaccination, and Diagnostics. Genes Immun. 2019, 20, 357–370. [Google Scholar] [CrossRef]
- Hinnebusch, B.J.; Erickson, D.L. Yersinia Pestis Biofilm in the Flea Vector and Its Role in the Transmission of Plague. Curr. Top. Microbiol. Immunol. 2008, 322, 229–248. [Google Scholar] [CrossRef]
- Tsang, T.M.; Felek, S.; Krukonis, E.S. Ail Binding to Fibronectin Facilitates Yersinia Pestis Binding to Host Cells and Yop Delivery. Infect. Immun. 2010, 78, 3358–3368. [Google Scholar] [CrossRef]
- Itoh, Y.; Rice, J.D.; Goller, C.; Pannuri, A.; Taylor, J.; Meisner, J.; Beveridge, T.J.; Preston, J.F.; Romeo, T. Roles of PgaABCD Genes in Synthesis, Modification, and Export of the Escherichia Coli Biofilm Adhesin Poly-Beta-1,6-N-Acetyl-D-Glucosamine. J. Bacteriol. 2008, 190, 3670–3680. [Google Scholar] [CrossRef]
- Pelludat, C.; Rakin, A.; Jacobi, C.A.; Schubert, S.; Heesemann, J. The Yersiniabactin Biosynthetic Gene Cluster of Yersinia Enterocolitica: Organization and Siderophore-Dependent Regulation. J. Bacteriol. 1998, 180, 538–546. [Google Scholar] [CrossRef]
- de Almeida, A.M.P.; Guiyoule, A.; Guilvout, I.; Iteman, I.; Baranton, G.; Carniel, E. Chromosomal Irp2 Gene in Yersinia: Distribution, Expression, Deletion and Impact on Virulence. Microb. Pathog. 1993, 14, 9–21. [Google Scholar] [CrossRef]
- Pechous, R.D.; Broberg, C.A.; Stasulli, N.M.; Miller, V.L.; Goldman, W.E. In Vivo Transcriptional Profiling of Yersinia Pestis Reveals a Novel Bacterial Mediator of Pulmonary Inflammation. mBio 2015, 6, e02302-14. [Google Scholar] [CrossRef]
- Almeida, A.M.P.d.; Guiyoule, A.; Leal, N.C.; Carniel, E. Survey of the Irp2 Gene among Yersinia Pestis Strains Isolated during Several Plague Outbreaks in Northeast Brazil. Mem. Inst. Oswaldo Cruz 1994, 89, 87–92. [Google Scholar] [CrossRef]
- Du, Y.; Rosqvist, R.; Forsberg, A. Role of Fraction 1 Antigen of Yersinia Pestis in Inhibition of Phagocytosis. Infect. Immun. 2002, 70, 1453–1460. [Google Scholar] [CrossRef] [PubMed]
- Leal-Balbino, T.; Leal, N.; Lopes, C.; Almeida, A. de Differences in the Stability of the Plasmids of Yersinia Pestis Cultures in Vitro: Impact on Virulence. Mem. Inst. Oswaldo Cruz 2004, 99, 727–732. [Google Scholar] [CrossRef][Green Version]
- Weening, E.H.; Cathelyn, J.S.; Kaufman, G.; Lawrenz, M.B.; Price, P.; Goldman, W.E.; Miller, V.L. The Dependence of the Yersinia Pestis Capsule on Pathogenesis Is Influenced by the Mouse Background. Infect. Immun. 2011, 79, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Anisimov, A.P.; Bakhteeva, I.V.; Panfertsev, E.A.; Svetoch, T.E.; Kravchenko, T.B.; Platonov, M.E.; Titareva, G.M.; Kombarova, T.I.; Ivanov, S.A.; Rakin, A.V.; et al. The Subcutaneous Inoculation of PH 6 Antigen Mutants of Yersinia Pestis Does Not Affect Virulence and Immune Response in Mice. J. Med. Microbiol. 2009, 58, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.D.; Weening, E.H.; Miner, T.A.; Miller, V.L. Temperature Control of PsaA Expression by PsaE and PsaF in Yersinia Pestis. J. Bacteriol. 2019, 201, e00217-19. [Google Scholar] [CrossRef] [PubMed]
- Hood, M.I.; Skaar, E.P. Nutritional Immunity: Transition Metals at the Pathogen–Host Interface. Nat. Rev. Microbiol. 2012, 10, 525–537. [Google Scholar] [CrossRef]
- Fetherston, J.D.; Kirillina, O.; Bobrov, A.G.; Paulley, J.T.; Perry, R.D. The Yersiniabactin Transport System Is Critical for the Pathogenesis of Bubonic and Pneumonic Plague. Infect. Immun. 2010, 78, 2045–2052. [Google Scholar] [CrossRef]
- Sebbane, F.; Lemaître, N.; Sturdevant, D.E.; Rebeil, R.; Virtaneva, K.; Porcella, S.F.; Hinnebusch, B.J. Adaptive Response of Yersinia Pestis to Extracellular Effectors of Innate Immunity during Bubonic Plague. Proc. Natl. Acad. Sci. USA 2006, 103, 11766–11771. [Google Scholar] [CrossRef]
- Zauberman, A.; Vagima, Y.; Tidhar, A.; Aftalion, M.; Gur, D.; Rotem, S.; Chitlaru, T.; Levy, Y.; Mamroud, E. Host Iron Nutritional Immunity Induced by a Live Yersinia Pestis Vaccine Strain Is Associated with Immediate Protection against Plague. Front. Cell Infect. Microbiol. 2017, 7, 277. [Google Scholar] [CrossRef]
- Yang, X.; Pan, J.; Wang, Y.; Shen, X. Type VI Secretion Systems Present New Insights on Pathogenic Yersinia. Front. Cell Infect. Microbiol. 2018, 8, 260. [Google Scholar] [CrossRef]
- Karimi, Y.; de Almeida, C.R.; de Almeida, A.M.; Keyvanfar, A.; Bourdin, M. Particularités Des Souches de Yersinia Pestis Isolées Dans Le Nord-Est Du Brésil. Ann. Microbiol. 1974, 125A, 243–246. [Google Scholar]

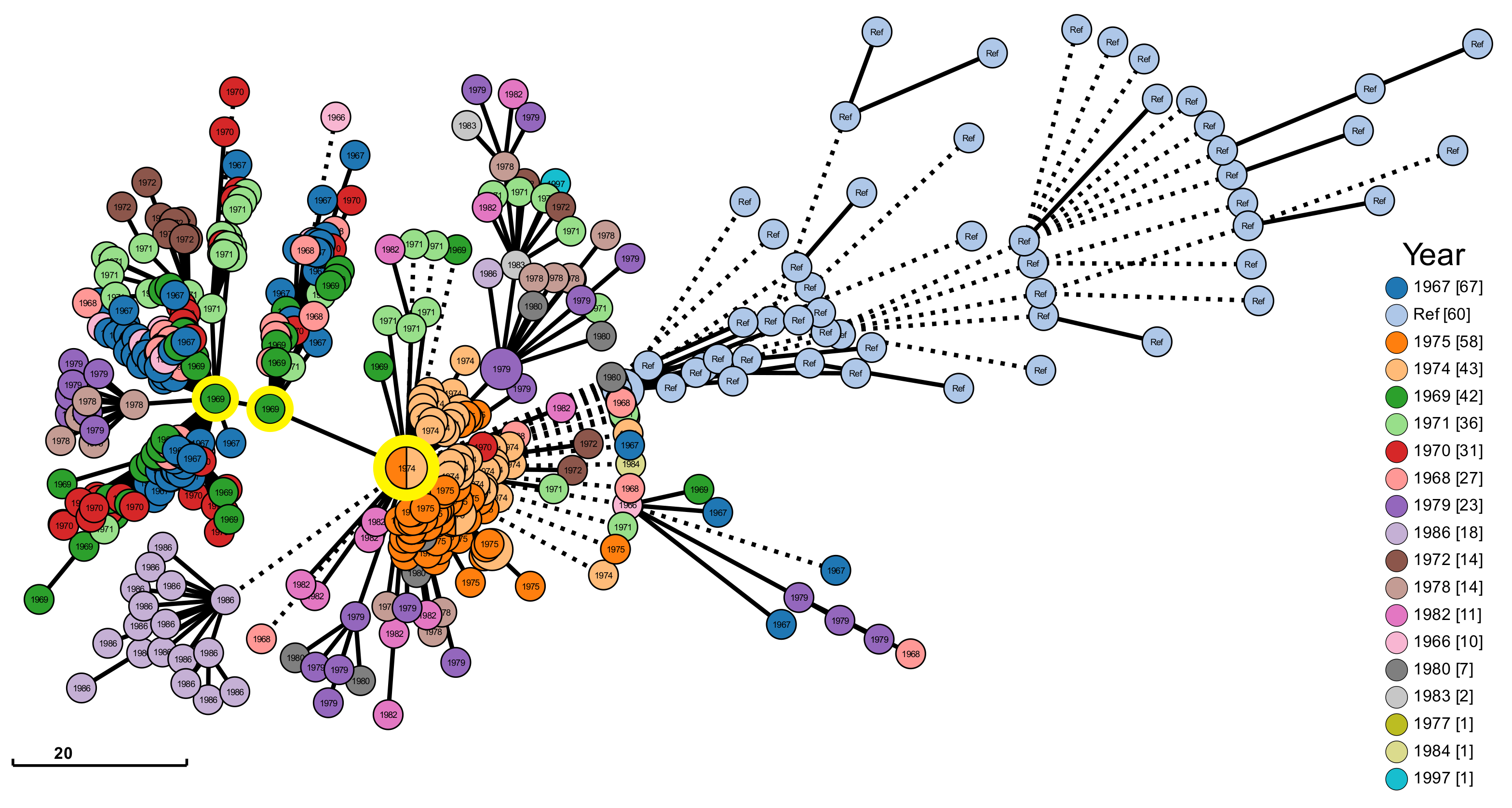
| Feature | Value |
|---|---|
| Average number of readings | 2183.187 |
| Average genome size | 4.6 Mb |
| Average N50 | 367,978 |
| Average depth | 42× |
| GC Content | 47% |
| Clade | Putative/Virulence Genes | Nonvirulence Genes |
|---|---|---|
| C | 10 | 43 |
| E | 13 | 0 |
| F | 0 | 4 |
| G | 25 | 41 |
| H | 9 | 83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pitta, J.L.d.L.P.; Bezerra, M.F.; Fernandes, D.L.R.d.S.; Block, T.d.; Novaes, A.d.S.; Almeida, A.M.P.d.; Rezende, A.M. Genomic Analysis of Yersinia pestis Strains from Brazil: Search for Virulence Factors and Association with Epidemiological Data. Pathogens 2023, 12, 991. https://doi.org/10.3390/pathogens12080991
Pitta JLdLP, Bezerra MF, Fernandes DLRdS, Block Td, Novaes AdS, Almeida AMPd, Rezende AM. Genomic Analysis of Yersinia pestis Strains from Brazil: Search for Virulence Factors and Association with Epidemiological Data. Pathogens. 2023; 12(8):991. https://doi.org/10.3390/pathogens12080991
Chicago/Turabian StylePitta, João Luiz de Lemos Padilha, Matheus Filgueira Bezerra, Diego Leandro Reis da Silva Fernandes, Tessa de Block, Ane de Souza Novaes, Alzira Maria Paiva de Almeida, and Antonio Mauro Rezende. 2023. "Genomic Analysis of Yersinia pestis Strains from Brazil: Search for Virulence Factors and Association with Epidemiological Data" Pathogens 12, no. 8: 991. https://doi.org/10.3390/pathogens12080991
APA StylePitta, J. L. d. L. P., Bezerra, M. F., Fernandes, D. L. R. d. S., Block, T. d., Novaes, A. d. S., Almeida, A. M. P. d., & Rezende, A. M. (2023). Genomic Analysis of Yersinia pestis Strains from Brazil: Search for Virulence Factors and Association with Epidemiological Data. Pathogens, 12(8), 991. https://doi.org/10.3390/pathogens12080991






