Clonal Diversity, Antibiotic Resistance, and Virulence Factor Prevalence of Community Associated Staphylococcus aureus in Southeastern Virginia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria
2.2. DNA Extraction
2.3. Whole-Genome Sequencing and Assembly
2.4. Sanger Sequencing and PCR
2.5. Statistics
3. Results
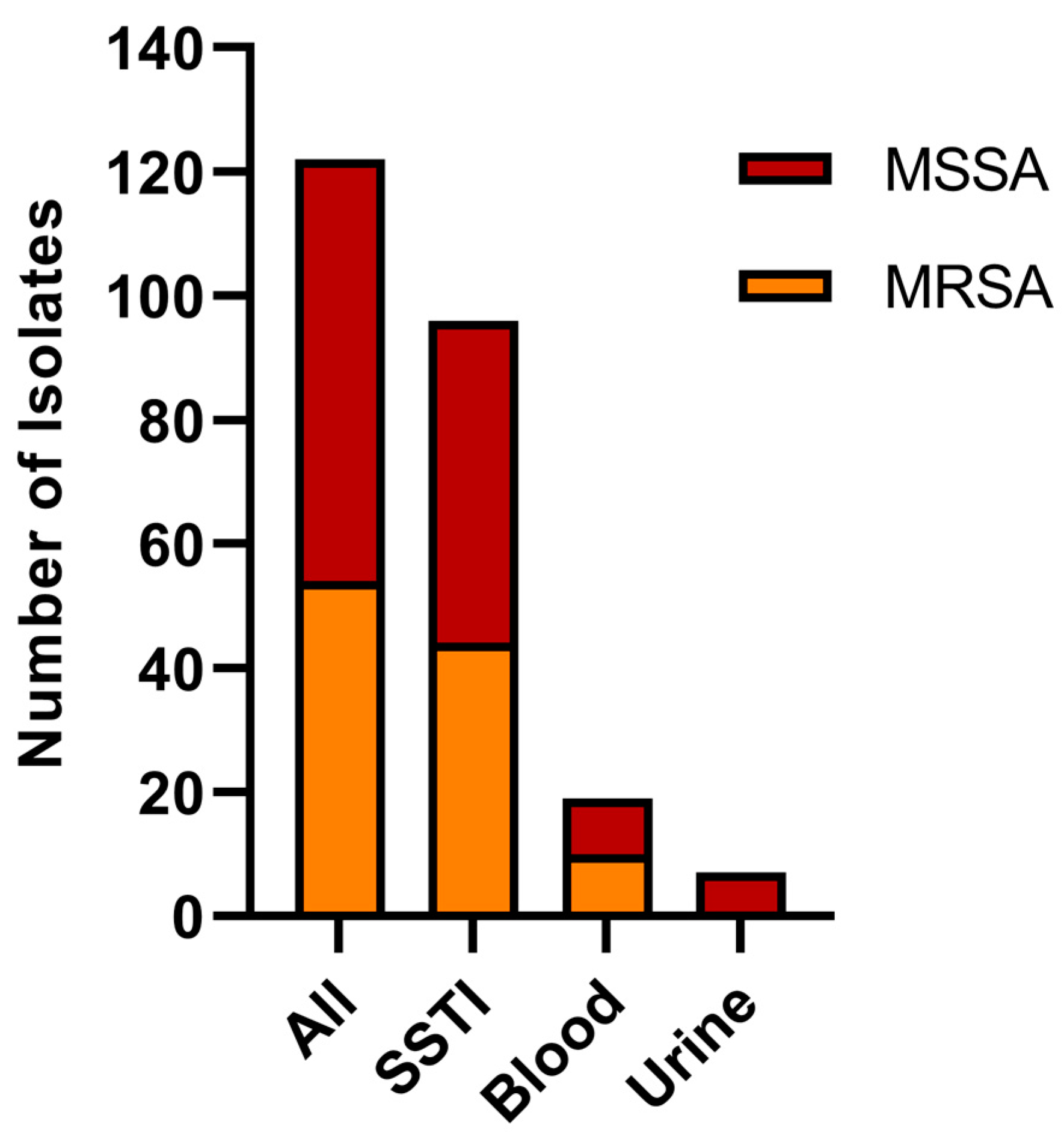
3.1. Sample Pool Composition
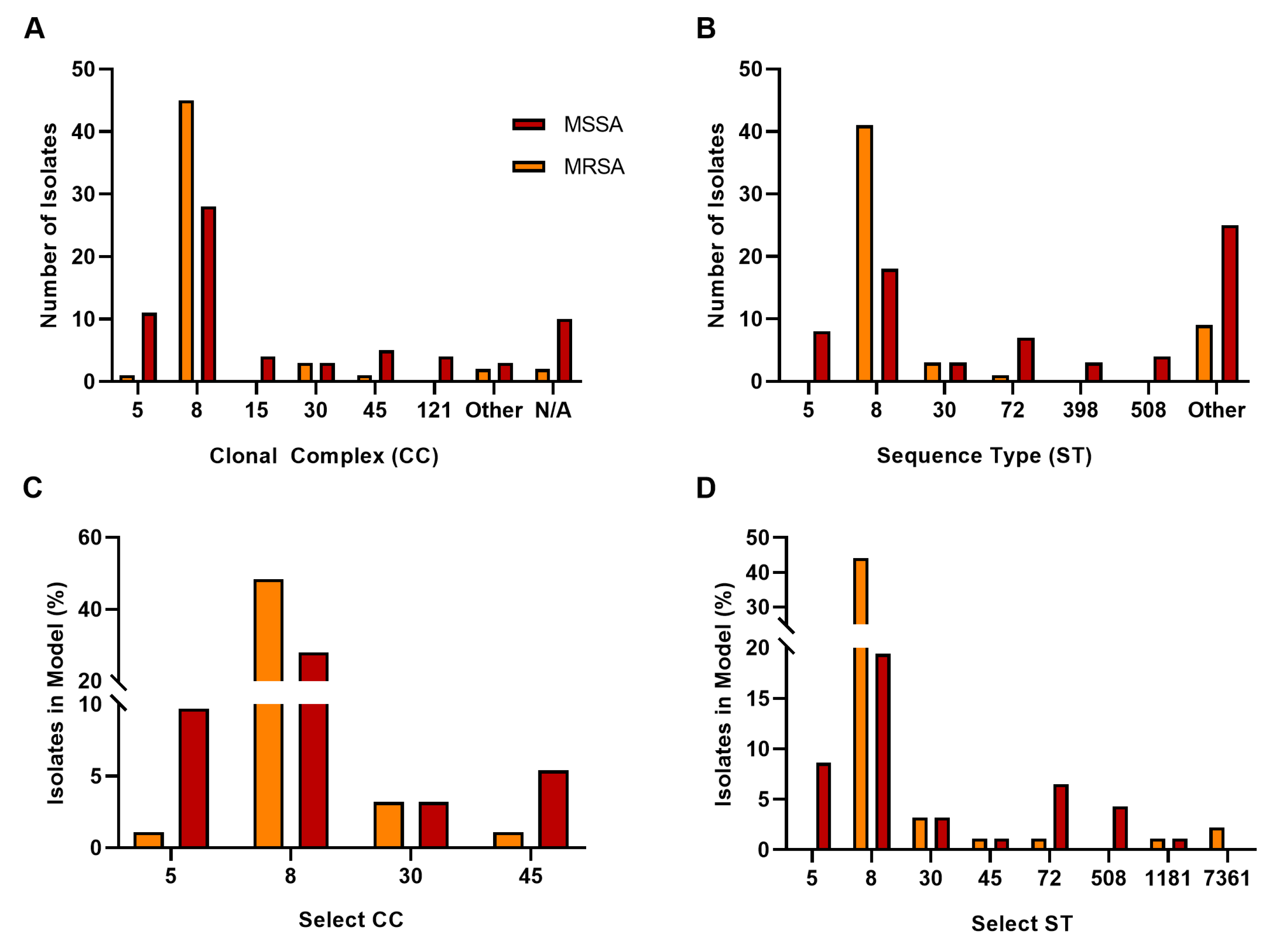
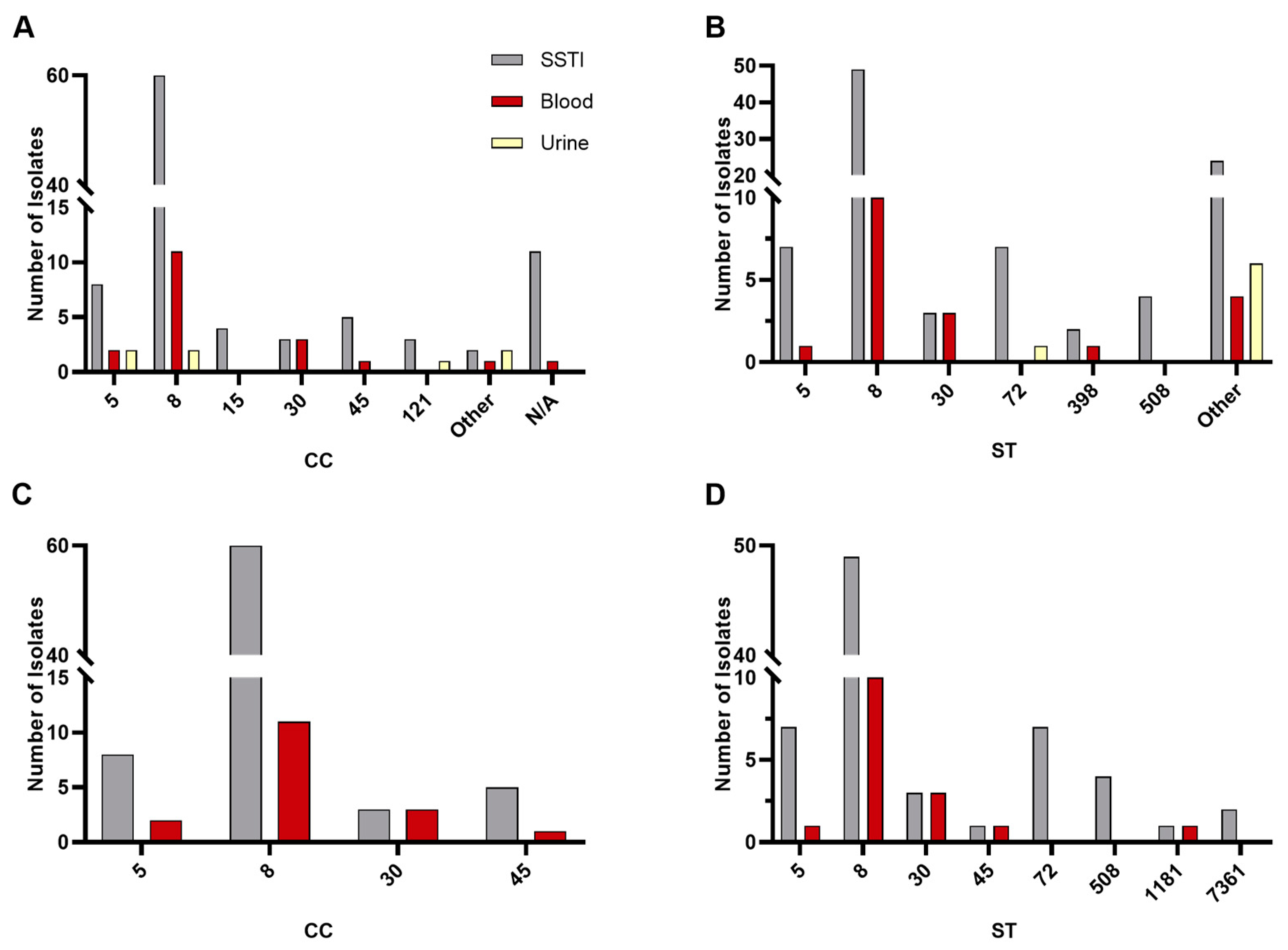
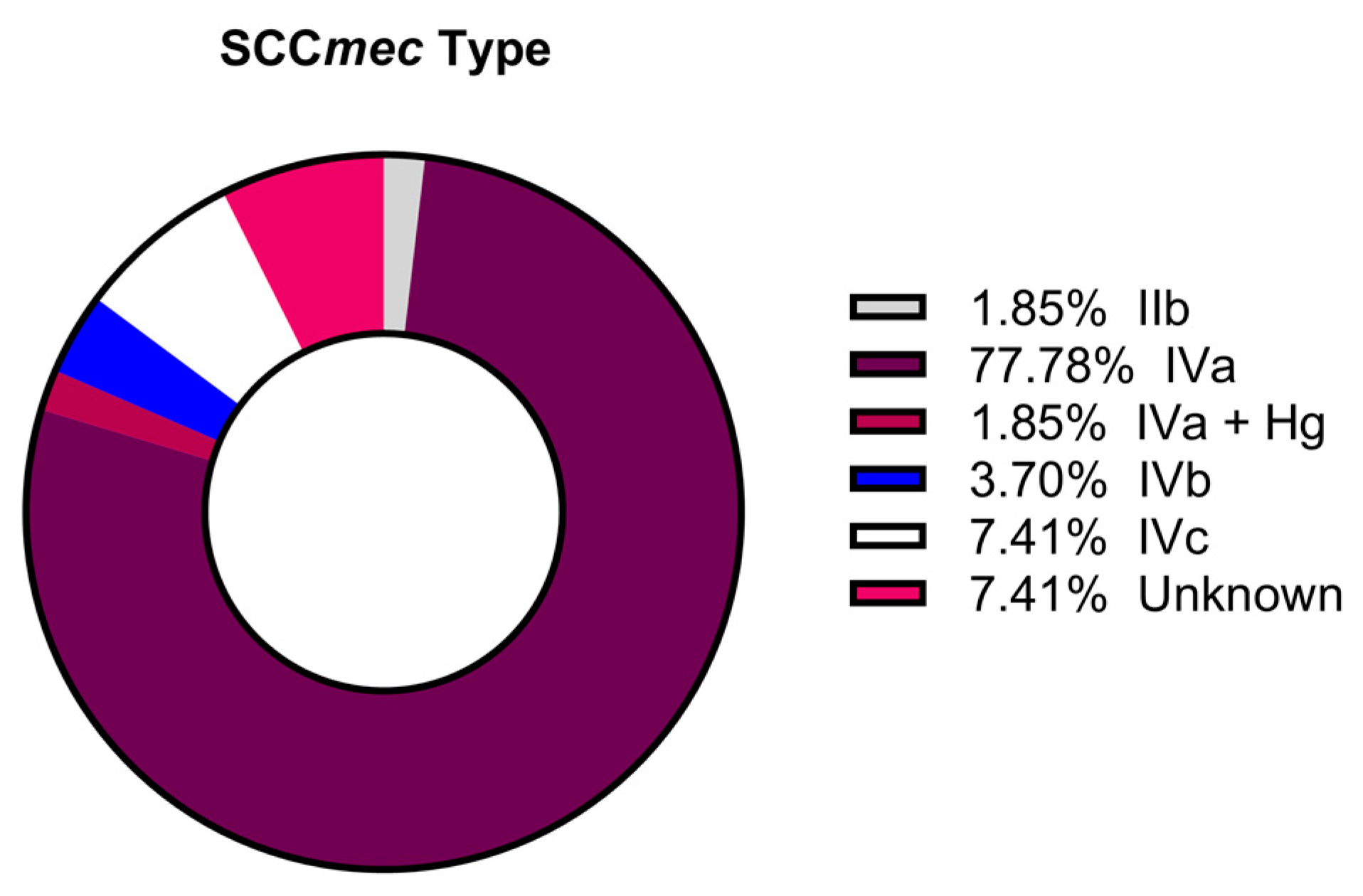
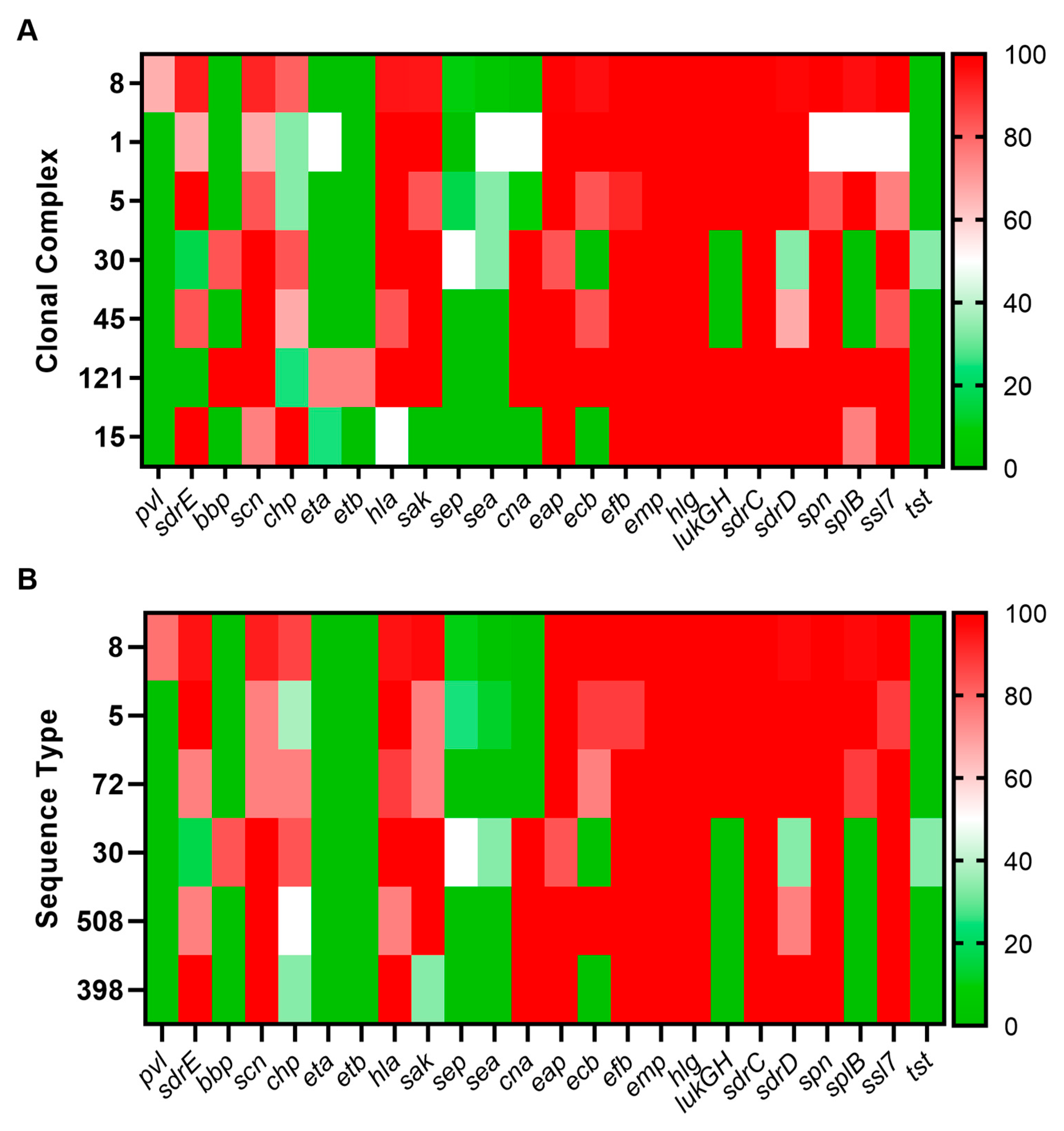
3.2. Sequence Type, Clonal Complex, and SCCmec Distribution
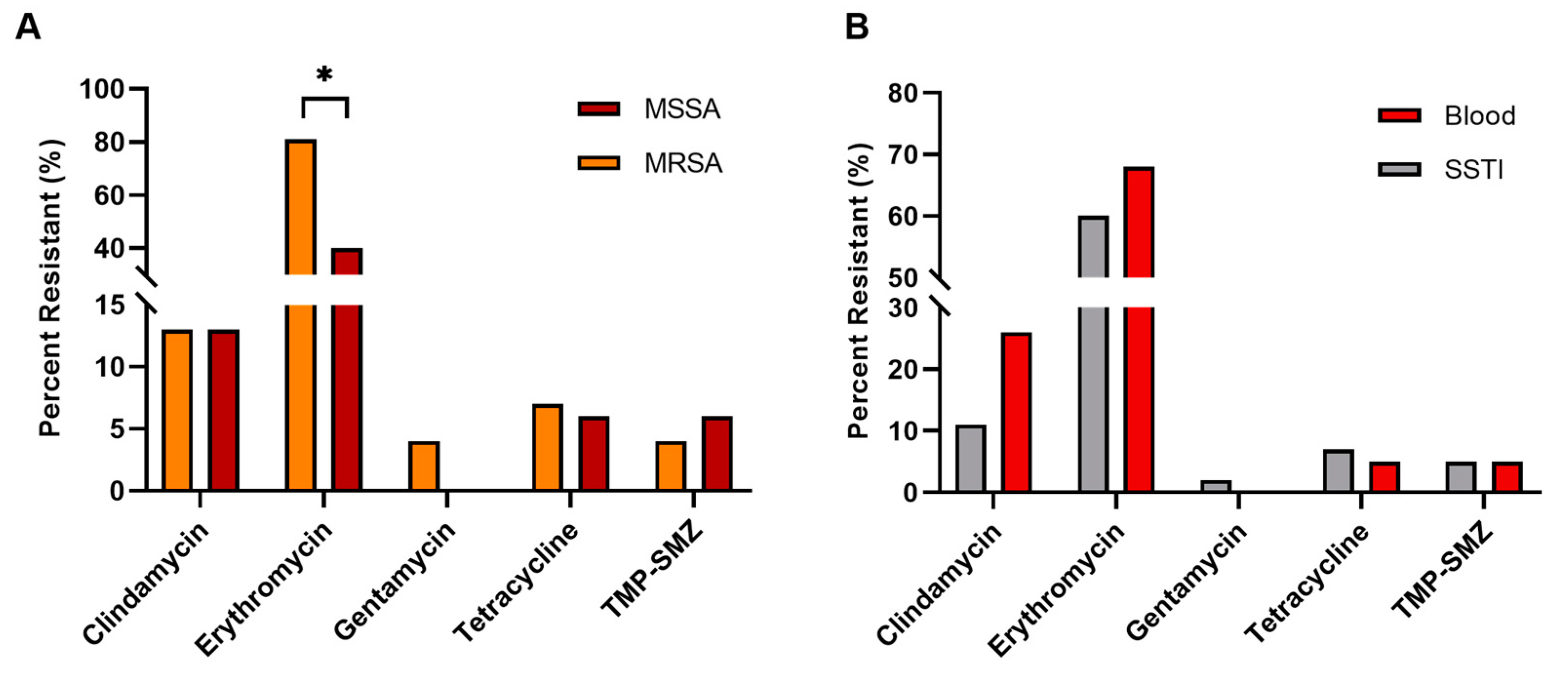
3.3. Antibiogram Data
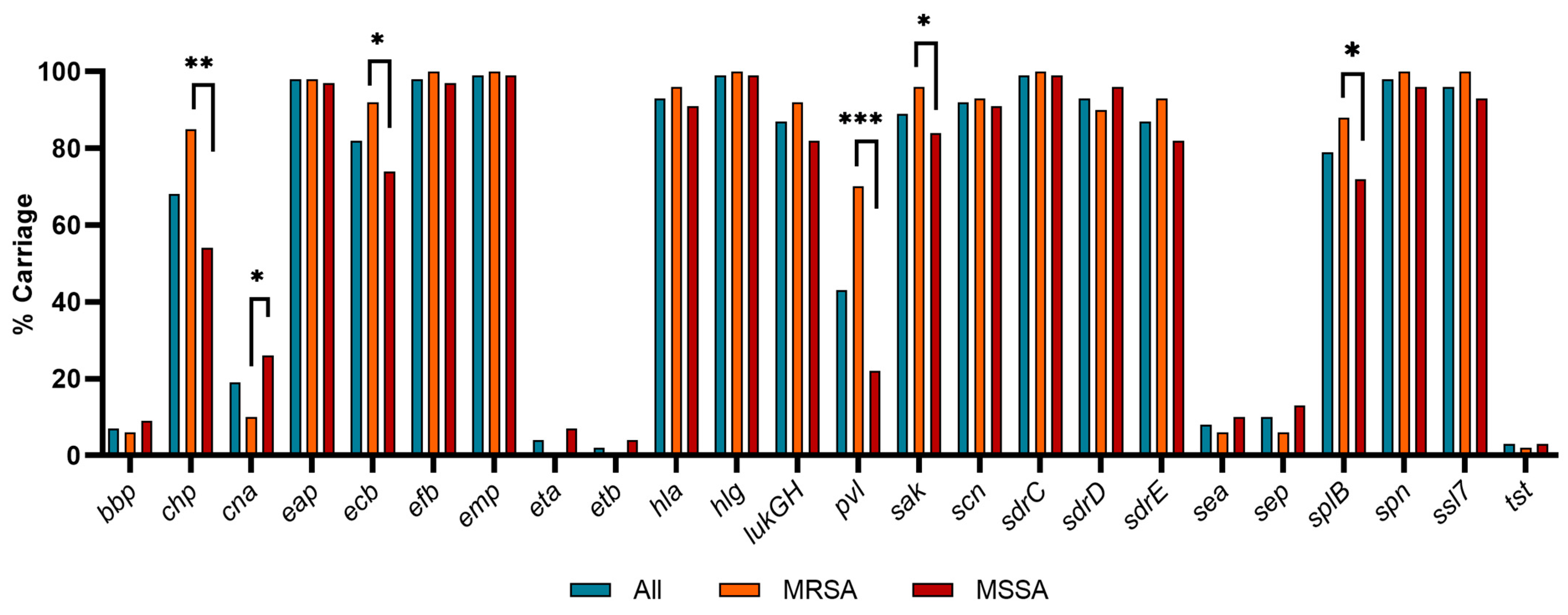
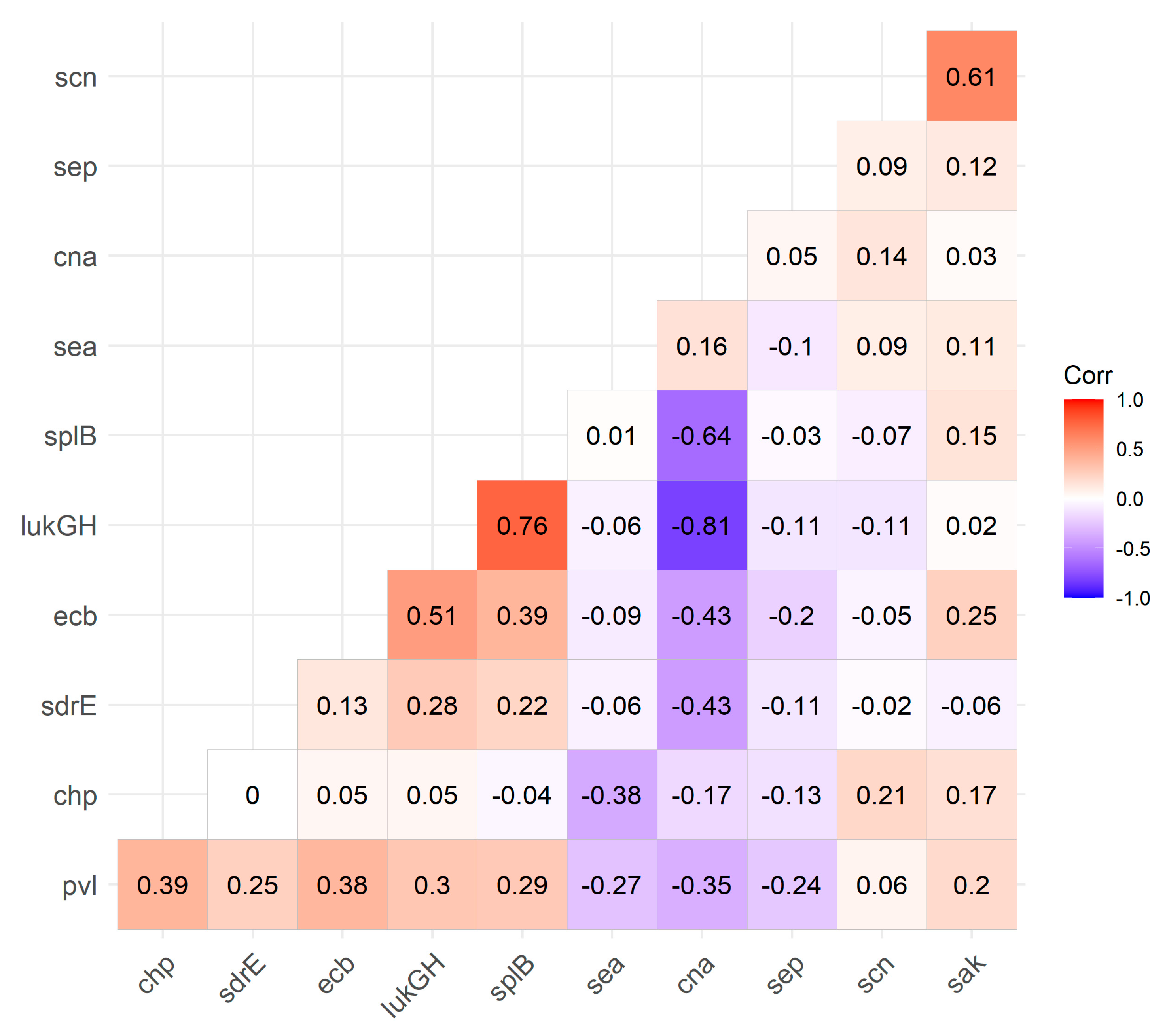
3.4. Virulence Factor Carriage
4. Discussion
4.1. Sequence Type, Clonal Complex, and SCCmec Distribution
4.2. Antibiograms
4.3. Virulence Factor Carriage
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Taylor, T.A.; Unakal, C.G. Staphylococcus aureus Infection; StatPearls Publishing: Treasure Island, FL, USA, 2023.
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Grigg, C.; Palms, D.; Stone, N.D.; Gualandi, N.; Bamberg, W.; Dumyati, G.; Harrison, L.H.; Lynfield, R.; Nadle, J.; Petit, S.; et al. Burden of Invasive Methicillin-Resistant Staphylococcus aureus Infections in Nursing Home Residents. J. Am. Geriatr. Soc. 2018, 66, 1581–1586. [Google Scholar] [CrossRef]
- Jones, M.; Jernigan, J.A.; Evans, M.E.; Roselle, G.A.; Hatfield, K.M.; Samore, M.H. Vital Signs: Trends in Staphylococcus aureus Infections in Veterans Affairs Medical Centers–United States, 2005–2017. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 220–224. [Google Scholar] [CrossRef]
- Koeck, M.; Como-Sabetti, K.; Boxrud, D.; Dobbins, G.; Glennen, A.; Anacker, M.; Jawahir, S.; See, I.; Lynfield, R. Burdens of Invasive Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus Disease, Minnesota, USA. Emerg. Infect. Dis. 2019, 25, 171–174. [Google Scholar] [CrossRef]
- Jackson, K.A.; Gokhale, R.H.; Nadle, J.; Ray, S.M.; Dumyati, G.; Schaffner, W.; Ham, D.C.; Magill, S.S.; Lynfield, R.; See, I. Public Health Importance of Invasive Methicillin-sensitive Staphylococcus aureus Infections: Surveillance in 8 US Counties, 2016. Clin. Infect Dis. 2020, 70, 1021–1028. [Google Scholar] [CrossRef]
- Hasman, H.; Moodley, A.; Guardabassi, L.; Stegger, M.; Skov, R.L.; Aarestrup, F.M. Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 2010, 141, 326–331. [Google Scholar] [CrossRef]
- Thapaliya, D.; Kadariya, J.; Capuano, M.; Rush, H.; Yee, C.; Oet, M.; Lohani, S.; Smith, T.C. Prevalence and Molecular Characterization of Staphylococcus aureus and Methicillin-resistant S. aureus on Children’s Playgrounds. Pediatr. Infect. Dis. J. 2019, 38, e43–e47. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G., Jr. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Diekema, D.J.; Pfaller, M.A.; Shortridge, D.; Zervos, M.; Jones, R.N. Twenty-Year Trends in Antimicrobial Susceptibilities Among Staphylococcus aureus From the SENTRY Antimicrobial Surveillance Program. Open Forum Infect. Dis. 2019, 6, S47–S53. [Google Scholar] [CrossRef]
- Siberry, G.K.; Tekle, T.; Carroll, K.; Dick, J. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin. Infect. Dis. 2003, 37, 1257–1260. [Google Scholar] [CrossRef]
- Walters, M.S.; Eggers, P.; Albrecht, V.; Travis, T.; Lonsway, D.; Hovan, G.; Taylor, D.; Rasheed, K.; Limbago, B.; Kallen, A. Vancomycin-Resistant Staphylococcus aureus–Delaware, 2015. MMWR Morb. Mortal. Wkly. Rep. 2015, 64, 1056. [Google Scholar] [CrossRef]
- Walters, M.S.; Rasheed, K.; Albrecht, V.; McAllister, S.; Limbago, B.; Kallen, A. Investigation and Control of Vancomycin-Resistant Staphylococcus aureus (VRSA): 2015 Update; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2015.
- Ghosh, S.; Banerjee, M. Methicillin resistance & inducible clindamycin resistance in Staphylococcus aureus. Indian J. Med. Res. 2016, 143, 362–364. [Google Scholar] [CrossRef]
- Jarajreh, D.; Aqel, A.; Alzoubi, H.; Al-Zereini, W. Prevalence of inducible clindamycin resistance in methicillin-resistant Staphylococcus aureus: The first study in Jordan. J. Infect. Dev. Ctries. 2017, 11, 350–354. [Google Scholar] [CrossRef]
- Rha, B.; See, I.; Dunham, L.; Kutty, P.K.; Moccia, L.; Apata, I.W.; Ahern, J.; Jung, S.; Li, R.; Nadle, J.; et al. Vital Signs: Health Disparities in Hemodialysis-Associated Staphylococcus aureus Bloodstream Infections - United States, 2017–2020. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 153–159. [Google Scholar] [CrossRef]
- Ricklin, D.; Tzekou, A.; Garcia, B.L.; Hammel, M.; McWhorter, W.J.; Sfyroera, G.; Wu, Y.Q.; Holers, V.M.; Herbert, A.P.; Barlow, P.N.; et al. A molecular insight into complement evasion by the staphylococcal complement inhibitor protein family. J. Immunol. 2009, 183, 2565–2574. [Google Scholar] [CrossRef]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol. Spectr. 2016, 4, 529–566. [Google Scholar] [CrossRef]
- Chavakis, T.; Wiechmann, K.; Preissner, K.T.; Herrmann, M. Staphylococcus aureus interactions with the endothelium: The role of bacterial “secretable expanded repertoire adhesive molecules” (SERAM) in disturbing host defense systems. Thromb. Haemost. 2005, 94, 278–285. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Simoes, M. Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef]
- Rooijakkers, S.H.; Ruyken, M.; Roos, A.; Daha, M.R.; Presanis, J.S.; Sim, R.B.; van Wamel, W.J.; van Kessel, K.P.; van Strijp, J.A. Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 2005, 6, 920–927. [Google Scholar] [CrossRef]
- de Haas, C.J.; Veldkamp, K.E.; Peschel, A.; Weerkamp, F.; Van Wamel, W.J.; Heezius, E.C.; Poppelier, M.J.; Van Kessel, K.P.; van Strijp, J.A. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J. Exp. Med. 2004, 199, 687–695. [Google Scholar] [CrossRef]
- Postma, B.; Poppelier, M.J.; van Galen, J.C.; Prossnitz, E.R.; van Strijp, J.A.; de Haas, C.J.; van Kessel, K.P. Chemotaxis inhibitory protein of Staphylococcus aureus binds specifically to the C5a and formylated peptide receptor. J. Immunol. 2004, 172, 6994–7001. [Google Scholar] [CrossRef] [PubMed]
- Loffler, B.; Hussain, M.; Grundmeier, M.; Bruck, M.; Holzinger, D.; Varga, G.; Roth, J.; Kahl, B.C.; Proctor, R.A.; Peters, G. Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophils. PLoS Pathog. 2010, 6, e1000715. [Google Scholar] [CrossRef] [PubMed]
- Hair, P.S.; Echague, C.G.; Sholl, A.M.; Watkins, J.A.; Geoghegan, J.A.; Foster, T.J.; Cunnion, K.M. Clumping factor A interaction with complement factor I increases C3b cleavage on the bacterial surface of Staphylococcus aureus and decreases complement-mediated phagocytosis. Infect. Immun. 2010, 78, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Sharp, J.A.; Echague, C.G.; Hair, P.S.; Ward, M.D.; Nyalwidhe, J.O.; Geoghegan, J.A.; Foster, T.J.; Cunnion, K.M. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS ONE 2012, 7, e38407. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.J.; Visai, L.; Kerrigan, S.W.; Speziale, P.; Foster, T.J. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect. Immun. 2011, 79, 3801–3809. [Google Scholar] [CrossRef] [PubMed]
- Ladhani, S. Understanding the mechanism of action of the exfoliative toxins of Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2003, 39, 181–189. [Google Scholar] [CrossRef]
- Kulhankova, K.; King, J.; Salgado-Pabon, W. Staphylococcal toxic shock syndrome: Superantigen-mediated enhancement of endotoxin shock and adaptive immune suppression. Immunol. Res. 2014, 59, 182–187. [Google Scholar] [CrossRef]
- Jiang, B.; Wang, Y.; Feng, Z.; Xu, L.; Tan, L.; Zhao, S.; Gong, Y.; Zhang, C.; Luo, X.; Li, S.; et al. Panton-Valentine leucocidin (PVL) as a potential indicator for prevalence, duration, and severity of Staphylococcus aureus osteomyelitis. Front. Microbiol. 2017, 8, 2355. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, T.; Zou, X.; Wu, S.; Zhu, J. Panton-Valentine leucocidin carrying Staphylococcus aureus causing necrotizing pneumonia inactivates the JAK/STAT signaling pathway and increases the expression of inflammatory cytokines. Infect. Genet. Evol. 2020, 86, 104582. [Google Scholar] [CrossRef]
- Castellazzi, M.L.; Bosis, S.; Borzani, I.; Tagliabue, C.; Pinzani, R.; Marchisio, P.; di Pietro, G.M. Panton-Valentine leukocidin Staphylococcus aureus severe infection in an infant: A case report and a review of the literature. Ital. J. Pediatr. 2021, 47, 158. [Google Scholar] [CrossRef]
- Wurster, J.I.; Bispo, P.J.M.; Van Tyne, D.; Cadorette, J.J.; Boody, R.; Gilmore, M.S. Staphylococcus aureus from ocular and otolaryngology infections are frequently resistant to clinically important antibiotics and are associated with lineages of community and hospital origins. PLoS ONE 2018, 13, e0208518. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Smith, D.; Turner, C.E.; Game, L.; Pichon, B.; Hope, R.; Hill, R.; Kearns, A.; Sriskandan, S. Clinical and molecular epidemiology of staphylococcal toxic shock syndrome in the United Kingdom. Emerg. Infect. Dis. 2018, 24, 258–266. [Google Scholar] [CrossRef]
- Ganesh, V.K.; Rivera, J.J.; Smeds, E.; Ko, Y.P.; Bowden, M.G.; Wann, E.R.; Gurusiddappa, S.; Fitzgerald, J.R.; Hook, M. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008, 4, e1000226. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Ko, Y.P.; Liang, X.; Ross, C.L.; Liu, Q.; Murray, B.E.; Hook, M. Collagen-binding microbial surface components recognizing adhesive matrix molecule (MSCRAMM) of Gram-positive bacteria inhibit complement activation via the classical pathway. J. Biol. Chem. 2013, 288, 20520–20531. [Google Scholar] [CrossRef] [PubMed]
- Eisenbeis, J.; Saffarzadeh, M.; Peisker, H.; Jung, P.; Thewes, N.; Preissner, K.T.; Herrmann, M.; Molle, V.; Geisbrecht, B.V.; Jacobs, K.; et al. The Staphylococcus aureus extracellular adherence protein Eap is a DNA binding protein capable of blocking neutrophil extracellular trap formation. Front. Cell. Infect. Microbiol. 2018, 8, 235. [Google Scholar] [CrossRef]
- Hammel, M.; Sfyroera, G.; Pyrpassopoulos, S.; Ricklin, D.; Ramyar, K.X.; Pop, M.; Jin, Z.; Lambris, J.D.; Geisbrecht, B.V. Characterization of Ehp, a secreted complement inhibitory protein from Staphylococcus aureus. J. Biol. Chem. 2007, 282, 30051–30061. [Google Scholar] [CrossRef]
- Amdahl, H.; Haapasalo, K.; Tan, L.; Meri, T.; Kuusela, P.I.; van Strijp, J.A.; Rooijakkers, S.; Jokiranta, T.S. Staphylococcal protein Ecb impairs complement receptor-1 mediated recognition of opsonized bacteria. PLoS ONE 2017, 12, e0172675. [Google Scholar] [CrossRef]
- Posner, M.G.; Upadhyay, A.; Abubaker, A.A.; Fortunato, T.M.; Vara, D.; Canobbio, I.; Bagby, S.; Pula, G. Extracellular fibrinogen-binding protein (Efb) from Staphylococcus aureus inhibits the formation of platelet-leukocyte complexes. J. Biol. Chem. 2016, 291, 2764–2776. [Google Scholar] [CrossRef]
- Koch, T.K.; Reuter, M.; Barthel, D.; Bohm, S.; van den Elsen, J.; Kraiczy, P.; Zipfel, P.F.; Skerka, C. Staphylococcus aureus proteins Sbi and Efb recruit human plasmin to degrade complement C3 and C3b. PLoS ONE 2012, 7, e47638. [Google Scholar] [CrossRef]
- Geraci, J.; Neubauer, S.; Pollath, C.; Hansen, U.; Rizzo, F.; Krafft, C.; Westermann, M.; Hussain, M.; Peters, G.; Pletz, M.W.; et al. The Staphylococcus aureus extracellular matrix protein (Emp) has a fibrous structure and binds to different extracellular matrices. Sci. Rep. 2017, 7, 13665. [Google Scholar] [CrossRef]
- Berube, B.J.; Bubeck Wardenburg, J. Staphylococcus aureus alpha-toxin: Nearly a century of intrigue. Toxins 2013, 5, 1140–1166. [Google Scholar] [CrossRef] [PubMed]
- Spaan, A.N.; van Strijp, J.A.G.; Torres, V.J. Leukocidins: Staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 2017, 15, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Rooijakkers, S.H.; van Wamel, W.J.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005, 7, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.J.; Corrigan, R.M.; van der Sluis, T.; Grundling, A.; Speziale, P.; Geoghegan, J.A.; Foster, T.J. The immune evasion protein Sbi of Staphylococcus aureus occurs both extracellularly and anchored to the cell envelope by binding lipoteichoic acid. Mol. Microbiol. 2012, 83, 789–804. [Google Scholar] [CrossRef]
- Jusko, M.; Potempa, J.; Kantyka, T.; Bielecka, E.; Miller, H.K.; Kalinska, M.; Dubin, G.; Garred, P.; Shaw, L.N.; Blom, A.M. Staphylococcal proteases aid in evasion of the human complement system. J. Innate Immun. 2014, 6, 31–46. [Google Scholar] [CrossRef]
- Feuillie, C.; Formosa-Dague, C.; Hays, L.M.; Vervaeck, O.; Derclaye, S.; Brennan, M.P.; Foster, T.J.; Geoghegan, J.A.; Dufrene, Y.F. Molecular interactions and inhibition of the staphylococcal biofilm-forming protein SdrC. Proc. Natl. Acad. Sci. USA 2017, 114, 3738–3743. [Google Scholar] [CrossRef]
- Askarian, F.; Ajayi, C.; Hanssen, A.M.; van Sorge, N.M.; Pettersen, I.; Diep, D.B.; Sollid, J.U.; Johannessen, M. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci. Rep. 2016, 6, 22134. [Google Scholar] [CrossRef]
- Cavallin, A.; Arozenius, H.; Kristensson, K.; Antonsson, P.; Otzen, D.E.; Bjork, P.; Forsberg, G. The spectral and thermodynamic properties of staphylococcal enterotoxin A, E, and variants suggest that structural modifications are important to control their function. J. Biol. Chem. 2000, 275, 1665–1672. [Google Scholar] [CrossRef]
- Omoe, K.; Hu, D.L.; Ono, H.K.; Shimizu, S.; Takahashi-Omoe, H.; Nakane, A.; Uchiyama, T.; Shinagawa, K.; Imanishi, K. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect. Immun. 2013, 81, 3627–3631. [Google Scholar] [CrossRef]
- Falugi, F.; Kim, H.K.; Missiakas, D.M.; Schneewind, O. Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 2013, 4, e00575-13. [Google Scholar] [CrossRef]
- Dasari, P.; Nordengrun, M.; Vilhena, C.; Steil, L.; Abdurrahman, G.; Surmann, K.; Dhople, V.; Lahrberg, J.; Bachert, C.; Skerka, C.; et al. The protease SplB of Staphylococcus aureus targets host complement components and inhibits complement-mediated bacterial opsonophagocytosis. J. Bacteriol. 2022, 204, e0018421. [Google Scholar] [CrossRef]
- de Jong, N.W.M.; Ramyar, K.X.; Guerra, F.E.; Nijland, R.; Fevre, C.; Voyich, J.M.; McCarthy, A.J.; Garcia, B.L.; van Kessel, K.P.M.; van Strijp, J.A.G.; et al. Immune evasion by a staphylococcal inhibitor of myeloperoxidase. Proc. Natl. Acad. Sci. USA 2017, 114, 9439–9444. [Google Scholar] [CrossRef]
- Bestebroer, J.; Aerts, P.C.; Rooijakkers, S.H.; Pandey, M.K.; Kohl, J.; van Strijp, J.A.; de Haas, C.J. Functional basis for complement evasion by staphylococcal superantigen-like 7. Cell. Microbiol. 2010, 12, 1506–1516. [Google Scholar] [CrossRef]
- Thomer, L.; Schneewind, O.; Missiakas, D. Multiple ligands of von Willebrand factor-binding protein (vWbp) promote Staphylococcus aureus clot formation in human plasma. J. Biol. Chem. 2013, 288, 28283–28292. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Enright, M.C.; Day, N.P.; Davies, C.E.; Peacock, S.J.; Spratt, B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000, 38, 1008–1015. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Conly, J.M. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mol. Cell. Probes 2012, 26, 218–221. [Google Scholar] [CrossRef]
- Tristan, A.; Ying, L.; Bes, M.; Etienne, J.; Vandenesch, F.; Lina, G. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 2003, 41, 4465–4467. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.R.; van Strijp, J.A.G.; Bagnoli, F.; Manetti, A.G.O. Virulence gene expression of Staphylococcus aureus in human skin. Front. Microbiol. 2021, 12, 692023. [Google Scholar] [CrossRef]
- Kato, F.; Kadomoto, N.; Iwamoto, Y.; Bunai, K.; Komatsuzawa, H.; Sugai, M. Regulatory mechanism for exfoliative toxin production in Staphylococcus aureus. Infect. Immun. 2011, 79, 1660–1670. [Google Scholar] [CrossRef]
- Karmakar, A.; Jana, D.; Dutta, K.; Dua, P.; Ghosh, C. Prevalence of Panton-Valentine leukocidin gene among community acquired Staphylococcus aureus: A real-time PCR study. J. Pathog. 2018, 2018, 4518541. [Google Scholar] [CrossRef]
- Oogai, Y.; Matsuo, M.; Hashimoto, M.; Kato, F.; Sugai, M.; Komatsuzawa, H. Expression of virulence factors by Staphylococcus aureus grown in serum. Appl. Environ. Microbiol. 2011, 77, 8097–8105. [Google Scholar] [CrossRef]
- Sabat, A.; Melles, D.C.; Martirosian, G.; Grundmann, H.; van Belkum, A.; Hryniewicz, W. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J. Clin. Microbiol. 2006, 44, 1135–1138. [Google Scholar] [CrossRef]
- Nielsen, L.N.; Roggenbuck, M.; Haaber, J.; Ifrah, D.; Ingmer, H. Diverse modulation of spa transcription by cell wall active antibiotics in Staphylococcus aureus. BMC Res. Notes 2012, 5, 457. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, Version R-4.2.3; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wickham, H.; Bryan, J. readxl: Read Excel Files. 2023. Available online: https://github.com/tidyverse/readxl (accessed on 23 October 2023).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D. dplyr: A Grammar of Data Manipulation. 2023. Available online: https://github.com/tidyverse/dplyr (accessed on 23 October 2023).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kassambara, A. ggcorrplot: Visualization of a Correlation Matrix Using ‘ggplot2’; Springer: New York, NY, USA, 2023. [Google Scholar]
- Mehta, C.R.; Patel, N.R. A hybrid algorithm for Fisher’s exact test in unordered RXC contingency tables. Commun. Stat. Theory Methods 1986, 15, 387–403. [Google Scholar] [CrossRef]
- O’Brien, L.; Kerrigan, S.W.; Kaw, G.; Hogan, M.; Penades, J.; Litt, D.; Fitzgerald, D.J.; Foster, T.J.; Cox, D. Multiple mechanisms for the activation of human platelet aggregation by Staphylococcus aureus: Roles for the clumping factors ClfA and ClfB, the serine-aspartate repeat protein SdrE and protein A. Mol. Microbiol. 2002, 44, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Ghasemian, A.; Najar Peerayeh, S.; Bakhshi, B.; Mirzaee, M. The microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) genes among clinical isolates of Staphylococcus aureus from hospitalized children. Iran. J. Pathol. 2015, 10, 258–264. [Google Scholar] [PubMed]
- Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Staphylococcal enterotoxins. Toxins 2010, 2, 2177–2197. [Google Scholar] [CrossRef] [PubMed]
- van Wamel, W.J.; Rooijakkers, S.H.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J. Bacteriol. 2006, 188, 1310–1315. [Google Scholar] [CrossRef]
- Sutter, D.E.; Milburn, E.; Chukwuma, U.; Dzialowy, N.; Maranich, A.M.; Hospenthal, D.R. Changing Susceptibility of Staphylococcus aureus in a US Pediatric Population. Pediatrics 2016, 137, e20153099. [Google Scholar] [CrossRef]
- Uehara, Y. Current Status of Staphylococcal Cassette Chromosome mec (SCCmec). Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef]
- Tenover, F.C.; Goering, R.V. Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J. Antimicrob. Chemother. 2009, 64, 441–446. [Google Scholar] [CrossRef]
- Diekema, D.J.; Richter, S.S.; Heilmann, K.P.; Dohrn, C.L.; Riahi, F.; Tendolkar, S.; McDanel, J.S.; Doern, G.V. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: Results from a nationwide surveillance study. Infect. Control Hosp. Epidemiol. 2014, 35, 285–292. [Google Scholar] [CrossRef]
- Sansom, S.E.; Benedict, E.; Thiede, S.N.; Hota, B.; Aroutcheva, A.; Payne, D.; Zawitz, C.; Snitkin, E.S.; Green, S.J.; Weinstein, R.A.; et al. Genomic update of phenotypic prediction rule for methicillin-resistant Staphylococcus aureus (MRSA) USA300 discloses jail transmission networks with increased resistance. Microbiol. Spectr. 2021, 9, e0037621. [Google Scholar] [CrossRef]
- Thiede, S.N.; Snitkin, E.S.; Trick, W.; Payne, D.; Aroutcheva, A.; Weinstein, R.A.; Popovich, K.J. Genomic epidemiology suggests community origins of healthcare-associated USA300 methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2022, 226, 157–166. [Google Scholar] [CrossRef]
- Valour, F.; Tasse, J.; Trouillet-Assant, S.; Rasigade, J.P.; Lamy, B.; Chanard, E.; Verhoeven, P.; Decousser, J.W.; Marchandin, H.; Bes, M.; et al. Methicillin-susceptible Staphylococcus aureus clonal complex 398: High prevalence and geographical heterogeneity in bone and joint infection and nasal carriage. Clin. Microbiol. Infect. 2014, 20, O772–O775. [Google Scholar] [CrossRef]
- Sharma-Kuinkel, B.K.; Mongodin, E.F.; Myers, J.R.; Vore, K.L.; Canfield, G.S.; Fraser, C.M.; Rude, T.H.; Fowler, V.G., Jr.; Gill, S.R. Potential influence of Staphylococcus aureus clonal complex 30 genotype and transcriptome on hematogenous infections. Open Forum Infect. Dis. 2015, 2, ofv093. [Google Scholar] [CrossRef] [PubMed]
- Bush, L.M.; Vazquez-Pertejo, M.T. Staphylococcal Infections. Merck Manual Professional Version. 2023. Available online: https://www.merckmanuals.com/professional/infectious-diseases/gram-positive-cocci/staphylococcal-infections (accessed on 25 October 2023).
- Calderwood, M.S.; Desjardins, C.A.; Sakoulas, G.; Nicol, R.; Dubois, A.; Delaney, M.L.; Kleinman, K.; Cosimi, L.A.; Feldgarden, M.; Onderdonk, A.B.; et al. Staphylococcal enterotoxin P predicts bacteremia in hospitalized patients colonized with methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2014, 209, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Kurt, K.; Rasigade, J.P.; Laurent, F.; Goering, R.V.; Zemlickova, H.; Machova, I.; Struelens, M.J.; Zautner, A.E.; Holtfreter, S.; Broker, B.; et al. Subpopulations of Staphylococcus aureus clonal complex 121 are associated with distinct clinical entities. PLoS ONE 2013, 8, e58155. [Google Scholar] [CrossRef]
- Shen, P.; Zhou, K.; Wang, Y.; Song, J.; Liu, Y.; Zhou, Y.; Xiao, Y. High prevalence of a globally disseminated hypervirulent clone, Staphylococcus aureus CC121, with reduced vancomycin susceptibility in community settings in China. J. Antimicrob. Chemother. 2019, 74, 2537–2543. [Google Scholar] [CrossRef]

| Gene | VF | Type 1 | Group 2 | Immune-Evasive | Ref. |
|---|---|---|---|---|---|
| bbp | Bone Sialoprotein Binding Protein | CW | MSCRAMM | Yes | [18] |
| clfA | Clumping Factor A | CW | MSCRAMM | Yes | [25,35] |
| chp | Chemotaxis Inhibitory Protein | S | Exoprotein | Yes | [22,23] |
| cna | Collagen Adhesin | CW | MSCRAMM | Yes | [36] |
| eap | Extracellular Adherence Protein | S | SERAM | Yes | [37] |
| ecb | Extracellular Complement Binding Protein | S | SERAM | Yes | [19,38,39] |
| efb | Extracellular Fibrinogen-Binding Protein | S | SERAM | Yes | [19,40,41] |
| emp | Extracellular Matrix Protein | S | SERAM | No | [42] |
| eta | Exfoliative Toxin A | S | Ex. Toxin | No | [28] |
| etb | Exfoliative Toxin B | S | Ex. Toxin | No | [28] |
| hla | α-hemolysin or Alpha Toxin | S | PF Toxin | Yes | [43] |
| hlg | γ-hemolysin or Gamma Toxin | S | PF Toxin | Yes | [44] |
| lukAB/GH | Leukocidin AB or Leukocidin GH | S | PF Toxin | Yes | [44] |
| pvl | Panton-Valentine Leukocidin | S | PF Toxin | Yes | [24] |
| sak | Staphylokinase | S | Protease | Yes | [45] |
| sbi | Staphylococcal Binder of Immunoglobulin | CW and S | Exoprotein | Yes | [27,41,46] |
| scn | Staphylococcal Complement Inhibitor | S | Exoprotein | Yes | [21] |
| scpA | Cysteine Protease Staphopain A | S | Protease | Yes | [47] |
| sdrC | Serine-Aspartate Repeat Protein C | CW | MSCRAMM | No | [48] |
| sdrD | Serine-Aspartate Repeat Protein D | CW | MSCRAMM | No | [49] |
| sdrE | Serine-Aspartate Repeat Protein E | CW | MSCRAMM | Yes | [18,26] |
| sea | Staphylococcal Enterotoxin A | S | Enterotoxin; Superantigen | Yes | [50] |
| sep | Staphylococcal Enterotoxin P | S | Enterotoxin | No | [51] |
| spA | Staphylococcal Protein A | CW and S | Exoprotein | Yes | [52] |
| splB | Serine Protease-Like Protein B | S | Protease | Yes | [53] |
| spn | Staphylococcal Peroxidase Inhibitor | S | Exoprotein | Yes | [54] |
| ssl7 | Staphylococcal Superantigen-Like 7 Protein | S | Exoprotein | Yes | [55] |
| tst | Toxic Shock Syndrome Toxin | S | Superantigen | Yes | [29] |
| vwb | von Willebrand Factor-Binding Protein | S | SERAM | Yes | [19,56] |
| Infection Type * | |||
|---|---|---|---|
| Antibiotic | Range (µg/mL) | Blood/SSTI | Urine |
| Clindamycin | 0.5–2 | X | N/A |
| Erythromycin | 0.5–4 | X | N/A |
| Gentamycin | 1–8 | X | X |
| Oxacillin | 0.25–2 | X | X |
| Penicillin G | 0.125–8 | X | X |
| Rifampin | 0.5–2 | X | X |
| Tetracycline | 0.5–8 | X | X |
| Trimethoprim- Sulfamethoxazole | 0.5/9.5–2/38 | X | X |
| Vancomycin | 0.5–16 | X | X |
| Nitrofurantoin | 16–64 | N/A | X |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Ref. |
|---|---|---|---|
| bbp | AACTACATCTAGTACTCAACAACAG | ATGTGCTTGAATAACACCATCATCT | [62] |
| clfA | ATTGGCGTGGCTTCAGTGCT | CGTTTCTTCCGTAGTTGCATTTG | [62] |
| chp | GGAATCAGTACACACCATCATTCAG | ATTTCTCAAACGTTCATCTAATTTTCC | [63] |
| etb | GTGGTAAAGGCGGACAACAT | TCAAATCGTTCCCCAAAGTG | [64] |
| hla | TATAGTCAGCTCAGTAACAACAACA | TGCATGCCATTTTCTTTATCATAAGTGAC | [63] |
| pvl | ATCATTAGGTAAAATGTCTGGACATGATCCA | GCATCAAGTGTATTGGATAGCAAAAGC | [65] |
| scn | GTTGATATTTTGCTTCTGACAT | AACGAAAAGTTAGCTAATGAAT | [66] |
| sdrE | AGAAAGTATACTGTAGGAACTG | GATGGTTTTGTAGTTACATCGT | [67] |
| spA | CAAACGGCACTACTGCTGAC | CATGGTTTGCTGGTTGCTTC | [68] |
| Category | n | Isolates |
|---|---|---|
| MRSA | 54 | 44% |
| MSSA | 68 | 56% |
| Blood | 19 | 15.6% |
| SSTI | 96 | 78.7% |
| Urine * | 7 | 5.8% |
| Blood MRSA | 10 | 8.2% |
| Blood MSSA | 9 | 7.4% |
| SSTI MRSA | 44 | 36% |
| SSTI MSSA | 52 | 42.6% |
| Gene | VF | Description of Action | Ref. |
|---|---|---|---|
| clfA | Clumping Factor A | Binds human fibrinogen, involved in biofilm formation and S. aureus-mediated platelet aggregation. Contributes to immune evasion by binding complement regulator Factor I. | [25,35,75] |
| scpA | Cysteine Protease Staphopain A | Protease with inhibitory effects on complement pathways. Impairs phagocytosis by neutrophils. | [47] |
| sbi | Staphylococcal Binder of Immunoglobulin | Binds IgG Fc; binds and activates host plasminogen to intefere with complement-mediated opsonization. | [27,41] |
| spA | Staphylococcal Protein A | Binds IgG Fc and cross-links the Fab domain of IgM to subvert opsonization and phagocytosis. | [52] |
| vwb | von Willebrand Factor-Binding Protein | Secreted adhesin that binds to plasma components and induces blood clots. Assists in strengthening abscess walls. | [56] |
| Gene | VF | Description of Action | Carriage (%) * | Ref. |
|---|---|---|---|---|
| chp | Chemotaxis Inhibitory Protein | Inhibits fMLP- and C5a-induced chemotaxis of neutrophils and monocytes. | All: 68; 85 R, 54 S; 74 SSTI, 47 B, 43 U | [22,23] |
| cna | Collagen Adhesin | Binds host collagen. Inhibits complement by binding the initiator protein C1q. | All: 19; 10 R, 26S | [36,76] |
| ecb | Extra-cellular Complement Binding Protein | Impairs complement-mediated phagocytosis by binding complement C3b or C3, and reduces the cofactor activity of CR1. | All: 82; 92 R, 74 S | [38,39] |
| pvl | Panton-Valentine Leukocidin | Bi-component leukocidin that forms β-barrel pores in host cells, with high specificity to human neutrophils. | All: 43; 70 R, 22 S; 52 SSTI, 16 B, 0 U | [24] |
| sak | Staphylokinase | Binds and activates host plasminogen to break down host extracellular matrices. Also removes IgG and C3b (opsonins) from the bacterial surface. | All: 89; 96 R, 84 S | [45] |
| splB | Serine Protease-Like Protein B | Cleaves and inactivates several complement components, inhibiting all three pathways, reducing bacterial killing via phagocytosis. | All: 79; 88 R, 72 S | [53] |
| sea | Staphylococcal Enterotoxin A | Commonly associated with food poisoning; causes emesis, diarrhea, and GI inflammation. Also known for nonspecific activation of T-cells, resulting in acute toxic shock. | All: 8; 4 SSTI, 4 B, 29 U | [50,77] |
| sep | Staphylococcal Enterotoxin P | Related to and often on the same pathogenicity island as sea, though produces much milder symptoms. | All: 10; 5 SSTI, 35 B, 14 U | [51,78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cranmer, K.D.; Pant, M.D.; Quesnel, S.; Sharp, J.A. Clonal Diversity, Antibiotic Resistance, and Virulence Factor Prevalence of Community Associated Staphylococcus aureus in Southeastern Virginia. Pathogens 2024, 13, 25. https://doi.org/10.3390/pathogens13010025
Cranmer KD, Pant MD, Quesnel S, Sharp JA. Clonal Diversity, Antibiotic Resistance, and Virulence Factor Prevalence of Community Associated Staphylococcus aureus in Southeastern Virginia. Pathogens. 2024; 13(1):25. https://doi.org/10.3390/pathogens13010025
Chicago/Turabian StyleCranmer, Katelyn D., Mohan D. Pant, Suzanne Quesnel, and Julia A. Sharp. 2024. "Clonal Diversity, Antibiotic Resistance, and Virulence Factor Prevalence of Community Associated Staphylococcus aureus in Southeastern Virginia" Pathogens 13, no. 1: 25. https://doi.org/10.3390/pathogens13010025
APA StyleCranmer, K. D., Pant, M. D., Quesnel, S., & Sharp, J. A. (2024). Clonal Diversity, Antibiotic Resistance, and Virulence Factor Prevalence of Community Associated Staphylococcus aureus in Southeastern Virginia. Pathogens, 13(1), 25. https://doi.org/10.3390/pathogens13010025








