Development of a One-Step Multiplex qPCR Assay for Detection of Methicillin and Vancomycin Drug Resistance Genes in Antibiotic-Resistant Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Isolates and Reference Strains
2.2. Antimicrobial Susceptibility Testing (AST)
2.3. Genomic DNA Extraction
2.4. Development of Primers and Hydrolysis Probes for mecA and vanA-vanB Antibotic Resistance Genes
2.5. One-Step Multiplex qPCR Assay
2.6. Analytical Sensitivity and Specificity
2.7. Sequencing Analysis
2.8. Statistical Analysis
3. Results
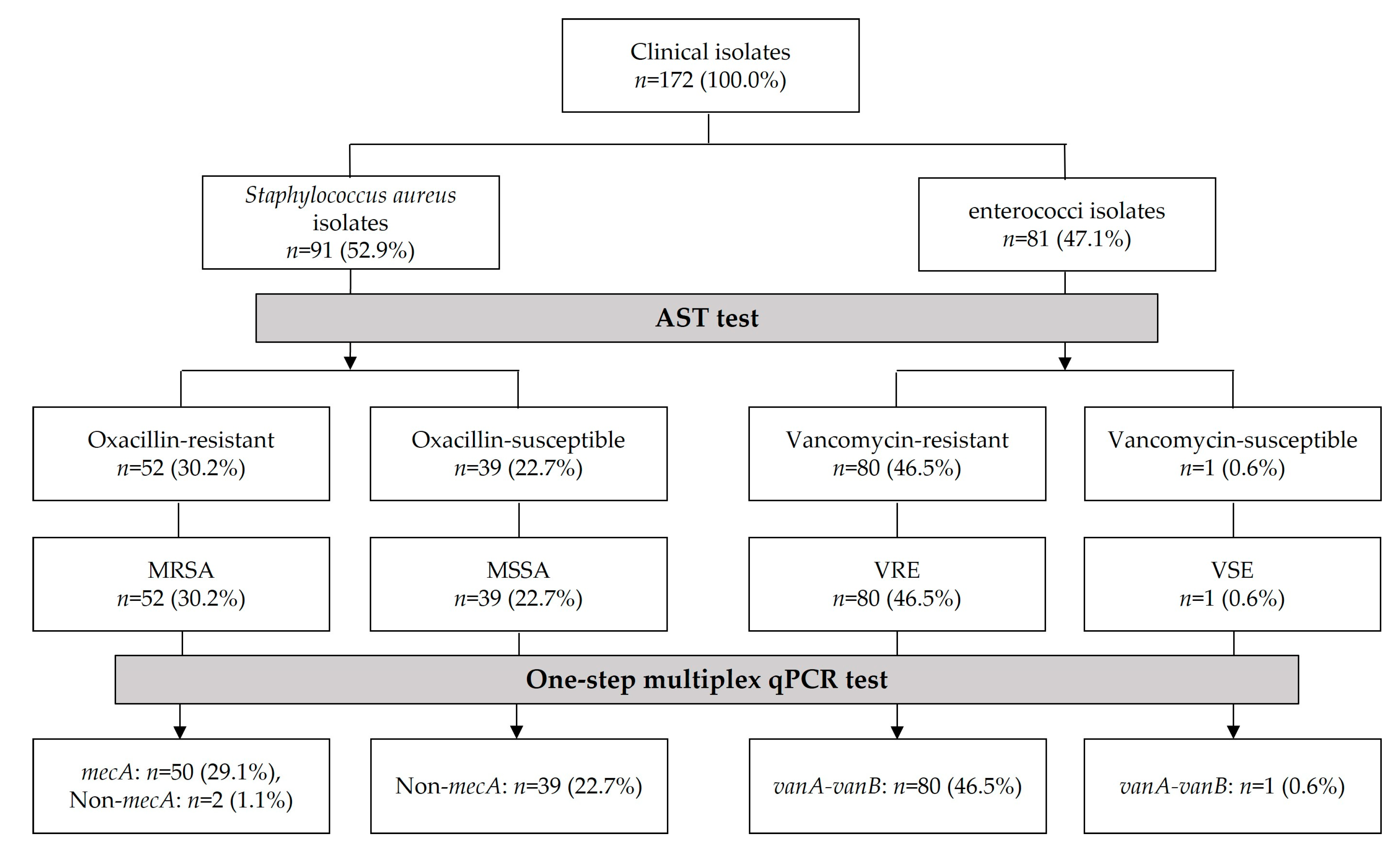
3.1. Antimicrobial Susceptibility Testing (AST) Data of Clinical Isolates
3.2. Analytical Sensitivity and Specificity of the One-Step Multiplex qPCR Assay
3.3. Identification of MRSA, MSSA, VRE, and VSE Isolates Using One-Step Multiplex qPCR Assay
3.4. Comparison of AST and One-Step Multiplex qPCR Assay for Clinical Isolates
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Park, J.E.; Kim, K.T.; Jang, D.H.; Song, Y.C.; Ha, N.J. Bacterial contamination and antimicrobial resistance of the surrounding environment influencing health. Korean J. Microbiol. 2014, 50, 101–107. [Google Scholar] [CrossRef]
- Kang, C.I.; Song, J.H. Antimicrobial resistance in Asia: Current epidemiology and clinical implications. Infect. Chemother. 2013, 45, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Choi, J.H.; Shin, D.W.; Kwon, K.T.; Kim, S.H.; Wi, Y.M.; Kwan, S.K. Conversion to colistin susceptibility by tigecycline exposure in colistin-resistant Klebsiella pneumoniae and Its implications to combination therapy. Int. J. Antimicrob. Agents 2024, 63, 107017. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Yoon, E.J.; Kim, D.K.; Jeong, S.H.; Shim, J.H.; Shin, J.H.; Shin, K.S.; Kim, Y.A.; Uh, Y.U.; Park, C.; et al. Establishment of the South Korean national antimicrobial resistance surveillance system, Kor-GLASS, in 2016. Perspective 2018, 23, 1700734. [Google Scholar] [CrossRef] [PubMed]
- Rho, K.H.; Jeong, H.R.; Kim, S.H.; Choi, H.J.; Jung, S.J.; Son, H.J.; Han, S.H.; Choi, J.Y.; Kim, S.W.; Kim, H.B.; et al. The Korean surgical site infection surveillance system report, 2018. Korean. J. Healthc. Assoc. Infect. Control Prev. 2020, 25, 128–136. [Google Scholar] [CrossRef]
- Snyder, G.M.; Thom, K.A.; Furuno, J.P.; Perencevich, E.N.; Roghmann, M.C.; Strauss, S.M.; Netzer, G.; Harris, A.D. Detection of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci by healthcare workers on infection control gown and gloves. Infect. Control Hosp. Epidemiol. 2008, 29, 583–589. [Google Scholar] [CrossRef]
- Nelwan, E.J.; Andayani, D.; Clarissa, G.; Pramada, T. Vancomycin-resistant Staphylococcus aureus infection post-liposuction in South Korea. Cureus 2021, 13, e14357. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Yoon, E.J.; Hong, J.S.; Choi, M.H.; Kim, H.S.; Kim, Y.R.; Kim, Y.A.; Uh, Y.; Shin, K.S.; Shin, J.H.; et al. Major bloodstream infection-causing bacterial pathogens and their antimicrobial resistance in South Korea, 2017–2019: Phase I Report from Kor-GLASS. Front. Microbiol. 2022, 12, 799084. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H.; Kim, C.J.; Choi, N.K.; Ahn, J.; Choe, P.G.; Park, W.B.; Kim, N.J.; Choi, H.J.; Bae, J.Y.; Kim, E.S.; et al. Clinical and economic burden of bacteremia due to multidrug-resistant organisms in Korea: A prospective case control study. Glob. Antimicrob. Resist. 2022, 31, 379–385. [Google Scholar] [CrossRef]
- Humphries, R.M. Update on Susceptibility Testing: Genotypic and Phenotypic Methods. Clin. Lab. Med. 2020, 40, 433–446. [Google Scholar]
- Gajic, I.; Kabic, J.; Kekic, D.; Jovicevic, M.; Milenkovic, M.; Mitic Culafic, D.; Trudic, A.; Ranin, L.; Opavski, N. Antimicrobial Susceptibility Testing: A Comprehensive Review of Currently Used Methods. Antibiotics 2022, 11, 427. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, M100, 32nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Chan, W.S.; Chan, T.M.; Lai, T.W.; Chan, J.F.; Lai, R.W.; Lai, C.K.; Tang, B.S. Complementary use of MALDI-TOF MS and real-time PCR–melt curve analysis for rapid identification of methicillin-resistant staphylococci and VRE. J. Antimicrob. Chemother. 2015, 70, 441–447. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Babiker, A.; Mustapha, M.M.; Pacey, M.P.; Shutt, K.A.; Ezeonwuka, C.D.; Ohm, S.L.; Cooper, V.S.; Marsh, J.W.; Doi, Y.; Harrison, L.H. Use of online tools for antimicrobial resistance prediction by whole-genome sequencing in methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). J. Glob. Antimicrob. Resist. 2019, 19, 136–143. [Google Scholar] [CrossRef]
- He, Y.H.; Ruan, G.J.; Hao, H.; Xue, F.; Ma, Y.K.; Zhu, S.N.; Zheng, B. Real-time PCR for the rapid detection of vanA, vanB and vanM genes. J. Microbiol. Immunol. Infect. 2020, 53, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Abdulsahib, S.S. Identification of the respiratory tract infection due to methicillin-resistant Staphylococcus aureu by TaqMan real-time PCR. Sciendo 2021, 9, 86–92. [Google Scholar]
- Costa, A.M.; Kay, I.; Palladino, S. Rapid detection of mecA and nuc genes in staphylococci by real-time multiplex polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2005, 51, 13–17. [Google Scholar] [CrossRef]
- Baek, Y.H.; Hong, S.B.; Shin, K.S. Simple and rapid detection of vancomycin-resistance gene from enterococci by loop-mediated isothermal amplification. Biomed. Sci. Lett. 2020, 26, 149–156. [Google Scholar] [CrossRef]
- Calfee, D.P. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci, and other gram-positives in healthcare. Curr. Opin. Infect. Dis. 2012, 25, 385–394. [Google Scholar] [CrossRef]
- Palmer, K.L.; Kos, V.N.; Gilmore, M.S. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr. Opin. Microbiol. 2010, 13, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Kriegeskorte, A.; Peters, G. Horizontal gene transfer boosts MRSA spreading. Nat. Med. 2012, 18, 662–663. [Google Scholar] [CrossRef] [PubMed]
- Cafini, F.; Thuy, N.T.L.; Román, F.; Prieto, J.; Dubrac, S.; Msadek, T.; Morikawa, K. Methodology for the study of horizontal gene transfer in Staphylococcus aureus. J. Vis. Exp. 2017, 121, e55087. [Google Scholar] [CrossRef]
- Moosavian, M.; Ghadri, H.; Zahra, S. Molecular detection of vanA and vanB genes among vancomycin-resistant enterococci in ICU-hospitalized patients in Ahvaz in Southwest of Iran. Infect. Drug Resist. 2018, 11, 2269–2275. [Google Scholar] [CrossRef] [PubMed]
- Wielders, C.L.C.; Fluit, A.C.; Brisse, S.; Verhoef, J.; Schmitz, F.J. mecA gene is widely disseminated in Staphylococcus aureus population. J. Clin. Microbiol. 2002, 40, 3970–3975. [Google Scholar] [CrossRef]
- Dhungel, S.; Rijal, K.R.; Yadav, B.; Dhungel, B.; Adhikari, N.; Shrestha, U.T.; Adhikari, B.; Banjara, M.R.; Ghimire, P. Methicillin-resistant Staphylococcus aureus (MRSA): Prevalence, antimicrobial susceptibility pattern, and detection of mecA gene among cardiac patients from a tertiary care heart center in Kathmandu, Nepal. Infect. Dis. Res. Treat. 2021, 14, 11786337211037355. [Google Scholar]
- Deurenberg, R.H.; Stobberingh, E.E. The evolution of Staphylococcus aureus. Infect. Genet. Evol. 2008, 8, 747–763. [Google Scholar] [CrossRef]
- Lee, H.W.; Yoon, J.H.; Sohn, J.H.; Lee, K.H.; Yeh, B.I.; Park, D.W.; Kim, H.W.; Choi, J.W. Detection of mecA gene in clinical isolates of Staphylococcus aureus by multiplex-PCR, and antimicrobial susceptibility of MRSA. J. Microbiol. Biotechnol. 2003, 13, 354–359. [Google Scholar]
- Seo, K.S.; Lim, J.Y.; Yoo, H.S.; Bae, W.K.; Park, Y.H. Comparison of vancomycin-resistant enterococci isolates from human, poultry and pigs in Korea. Vet. Microbiol. 2005, 106, 225–233. [Google Scholar] [CrossRef]
- Chang, E.; Kim, H.B. It is time to address the isolation policy for patients colonized with vancomycin-resistant enterococci. Korean J. Healthc. Assoc. Infect. Control Prev. 2021, 26, 16–23. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin resistance in gram-positive cocci. Clin. Infect. Dis. 2006, 42, S25–S34. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.G. Resistance mechanism and epidemiology of vancomycin-resistant enterococci. Korean J. Clin. Microbiol. 2008, 11, 71–77. [Google Scholar] [CrossRef][Green Version]
- Arredondo-Alonso, S.; Top, J.; Corander, J.; Willems, R.J.L.; Schürch, A.C. Mode and dynamics of vanA-type vancomycin resistance dissemination in Dutch hospitals. Genome Med. 2021, 13, 9. [Google Scholar] [CrossRef]
- Mirzaie, S.; Faghiri, I.; Badouei, M.A.; Madani, S.A. Molecular detection and occurrence of vancomycin resistance genes (van A, B, C1, C2/C3) among Enterococcus species isolated from farm ostriches. Vet. Med. Sci. 2023, 9, 226–233. [Google Scholar] [CrossRef]
- Guzman Prieto, A.M.; van Schaik, W.; Rogers, M.R.; Coque, T.M.; Baquero, F.; Corander, J.; Willems, R.J. Global Emergence and Dissemination of Enterococci as Nosocomial Pathogens: Attack of the Clones? Front. Microbiol. 2016, 26, 788. [Google Scholar] [CrossRef]
- van Belkum, A.; Burnham, C.A.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef]
- Gunson, R.N.; Bennett, S.; Maclean, A.; Carman, W.F. Using multiplex real time PCR in order to streamline a routine diagnostic service. J. Clin. Virol. 2008, 43, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Lee, S.Y.; Seo, S.H.; Lee, J.; Moon, D.C.; Yoo, J.S.; Choi, Y.H. Antimicrobial Resistance of Major Clinical Pathogens Isolated from General Hospitals in Korea: Results from the 2nd Phase (2020–2022) Kor-GLASS. Public Health Wkly. Rep. 2024, 17, 1034–1054. [Google Scholar]
- Maurin, M. Real-time PCR as a diagnostic tool for bacterial diseases. Expert Rev. Mol. Diagn. 2012, 12, 731–754. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.A.; Kumari, M.; Iyengar, S. Method for improving the quality of genomic DNA obtained from minute quantities of tissue and blood samples using Chelex 100 resin. Biol. Proced. Online 2018, 20, 12. [Google Scholar] [CrossRef]
- Hwang, I.; Kim, S.; Jung, T.; Kim, Y.; Kim, S. DNA Mutation Pattern of gyrA and gyrB Genes according to the SCCmec Subtype of Quinolone-resistant Staphylococcus aureus Isolates from Blood Culture. Korean J. Clin. Lab. Sci. (KJCLS) 2024, 56, 115–124. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Cho, E. National Infectious Diseases Surveillance data of South Korea. Epidemiol. Health 2014, 36, e2014030. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Bai, X.; Li, W.; Gao, X.; Zhang, F.; Sun, Z.; Zhang, H. Design of Primers for Evaluation of Lactic Acid Bacteria Populations in Complex Biological Samples. Front. Microbiol. 2018, 9, 2045. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Berger, P.; Raoult, D. Systematic 16S rRNA Gene Sequencing of Atypical Clinical Isolates Identified 27 New Bacterial Species Associated with Humans. J. Clin. Microbiol. 2004, 42, 2197–2202. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Patel, R. Molecular diagnostics for genotypic detection of antibiotic resistance: Current landscape and future directions. JAC Antimicrob. Resist. 2023, 5, dlad018. [Google Scholar] [CrossRef] [PubMed]
- Hasanpour, A.H.; Sepidarkish, M.; Mollalo, A.; Ardekani, A.; Almukhtar, M.; Mechaal, A.; Hosseini, S.R.; Bayani, M.; Javanian, M.; Rostami, A. The global prevalence of methicillin-resistant Staphylococcus aureus colonization in residents of elderly care centers: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2023, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Werner, G. Antibiotic susceptibility testing for therapy and antimicrobial resistance surveillance: Genotype beats phenotype? Future Microbiol. 2022, 17, 1093–1097. [Google Scholar] [CrossRef]
- Qodrati, M.; SeyedAlinaghi, S.; Dehghan Manshadi, S.A.; Abdollahi, A.; Dadras, O. Antimicrobial susceptibility testing of Staphylococcus aureus isolates from patients at a tertiary hospital in Tehran, Iran, 2018–2019. Eur. J. Med. Res. 2022, 27, 152. [Google Scholar] [CrossRef]
- Wang, W.Y.; Chen, Y.H.; Lee, Y.L.; Chiu, C.F.; Tsao, S.M. Comparative analysis of two commercial automated systems with agar dilution for oxacillin susceptibility and their association with genotypes of invasive Staphylococcus aureus isolates (2011–2021). Infect. Drug Resist. 2024, 17, 1121–1129. [Google Scholar] [CrossRef]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—A review of recent developments in MRSA management and treatment. Crit. Care 2017, 21, 211. [Google Scholar] [CrossRef] [PubMed]
- Rios, R.; Reyes, J.; Carvajal, L.P.; Rincon, S.; Panesso, D.; Echeverri, A.M.; Dinh, A.; Kolokotronis, S.O.; Narechania, A.; Tran, T.T.; et al. Genomic epidemiology of vancomycin-resistant Enterococcus faecium (VREfm) in Latin America: Revisiting the global VRE population structure. Sci. Rep. 2020, 10, 5636. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.S.; Jung, Y.H.; Kim, H.S.; Lee, Y.S.; Park, C.; Lee, K.J.; Cha, J.O. Prevalence of major methicillin-resistant Staphylococcus aureus clones in Korea between 2001 and 2008. Ann. Lab. Med. 2016, 36, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Song, K.H. Antibiotics for multidrug-resistant gram-positive bacteria. J. Korean Med. Assoc. 2022, 65, 478–489. [Google Scholar] [CrossRef]
- Kim, D.; Jeong, S.H. Current status of multidrug-resistant bacteria. J. Korean Med. Assoc. 2022, 65, 468–477. [Google Scholar] [CrossRef]
- Kim, D.; Choi, M.H.; Hong, J.S.; Shin, J.H.; Jeong, S.H. Current status and prospects of the national antimicrobial resistance surveillance system, Kor-GLASS. Korean J. Healthc. Assoc. Infect. Control Prev. 2022, 27, 96–103. [Google Scholar] [CrossRef]
- Korea Disease Control and Prevention Agency. Korea Home Page. Available online: https://www.korea.kr/archive/expDocView.do?docId=40204&group=S&pageIndex=2&pageUnit=20&startDate=2021-11-23&endDate=2022-11-23&srchKeyword=&srchCode=&codeLevel1=&codeLevel2=&allChkYN=&period= (accessed on 4 September 2024).
- McLain, J.E.; Cytryn, E.; Durso, L.M.; Young, S. Culture-based methods for detection of antibiotic resistance in agroecosystems: Advantages, challenges, and gaps in knowledge. J. Environ. Qual. 2016, 45, 432–440. [Google Scholar] [CrossRef]
- Sanchini, A. Recent Developments in Phenotypic and Molecular Diagnostic Methods for Antimicrobial Resistance Detection in Staphylococcus aureus: A Narrative Review. Diagnostics 2022, 12, 208. [Google Scholar] [CrossRef] [PubMed]
- Tajeddin, E.; Rashidan, M.; Razaghi, M.; Javadi, S.S.S.; Sherafat, S.J.; Alebouyeh, M.; Sarbazi, M.R.; Mansouri, N.; Zali, M.R. The role of the intensive care unit environment and health-care workers in the transmission of bacteria associated with hospital acquired infections. J. Infect. Public Health 2016, 9, 13–23. [Google Scholar] [CrossRef]
- Kim, H.J.; Na, S.W.; Alodaini, H.A.; Al-Dosary, M.A.; Nandhakumari, P.; Dyona, L. Prevalence of multidrug-resistant bacteria associated with polymicrobial infections. J. Infect. Public Health 2021, 14, 1864–1869. [Google Scholar] [CrossRef]
- Zautner, A.E.; Groß, U.; Emele, M.F.; Hagen, R.M.; Frickmann, H. More pathogenicity or just more pathogens?—on the interpretation problem of multiple pathogen detections with diagnostic multiplex assays. Front. Microbiol. 2017, 8, 1210. [Google Scholar] [CrossRef] [PubMed]
- Zirakzadeh, A.; Patel, R. Vancomycin-resistant enterococci: Colonization, infection, detection, and treatment. In Mayo Clinic Proceedings; Elsevier: Amsterdam, The Netherlands, 2006; Volume 81, pp. 529–536. [Google Scholar]
- Kim, M.J.; Kang, D.H.; Park, J.I.; Choi, T.Y. Evaluation of ChromID MRSA for the detection of methicillin-resistant Staphylococcus aureus. Korean J. Clin. Microbiol. 2009, 12, 169–173. [Google Scholar] [CrossRef][Green Version]
- van Hal, S.J.; Stark, D.; Lockwood, B.; Marriott, D.; Harkness, J. Methicillin-resistant Staphylococcus aureus (MRSA) detection: Comparison of two molecular methods (IDI-MRSA PCR assay and genotype MRSA direct PCR assay) with three selective MRSA agars (MRSA ID, MRSA elect, and CHROMagar MRSA) for use with infection-control swabs. J. Clin. Microbiol. 2009, 47, 1818–1823. [Google Scholar] [PubMed]
- Kim, H.S.; Park, S.B.; Kim, S.H.; Kim, S.H.; Hyun, S.H.; Kim, Y.K. Molecular genetic characteristics of methicillin-resistant Staphylococcus aureus isolated from patients and environment of general hospital intensive care unit in a Chungnam province, Korea. Korean J. Clin. Lab. Sci. 2018, 50, 110–117. [Google Scholar] [CrossRef]
- Kurkela, S.; Brown, D.W.G. Molecular diagnostic techniques. Medicine 2009, 37, 535–540. [Google Scholar] [CrossRef] [PubMed]
- March-Rosselló, G.A. Rapid methods for detection of bacterial resistance to antibiotics. Enferm. Infecc. Microbiol. Clin. 2017, 35, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Yamin, D.; Uskoković, V.; Wakil, A.M.; Goni, M.D.; Shamsuddin, S.H.; Mustafa, F.H.; Alfouzan, W.A.; Alissa, M.; Alshengeti, A.; Almaghrabi, R.H.; et al. Current and future technologies for the detection of antibiotic-resistant bacteria. Diagnostics 2023, 13, 3246. [Google Scholar] [CrossRef]
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern tools for rapid diagnostics of antimicrobial resistance. Front. Cell. Infect. Microbiol. 2020, 10, 308. [Google Scholar] [CrossRef]
- Kwak, O.S.; Kwon, M.H.; Jeong, J.H.; Kang, M.I.; Cheun, J.Y.; Lee, G.E.; Kim, Y.K.; Choi, E.G.; Na, M.J.; Kwon, H.U.; et al. Molecular Epidemiology and Antimicrobial Resistance of Methicillin-resistant Staphylococcus aureus Isolated from Nasal Swab at Intensive Care Unit. Tuberc. Respir. Dis. 2008, 65, 91–98. [Google Scholar] [CrossRef]
- Giurazza, R.; Mazza, M.C.; Andini, R.; Sansone, P.; Pace, M.C.; Durante-Mangoni, E. Emerging treatment options for multi-drug-resistant bacterial infections. Life 2021, 11, 519. [Google Scholar] [CrossRef]
- van den Brand, M.; Peters, R.P.H.; Catsburg, A.; Rubenjan, A.; Broeke, F.J.; van den Dungen, F.A.M.; van Weissenbruch, M.M.; van Furth, A.M.; Kõressaar, T.; Remm, M.; et al. Development of a multiplex real-time PCR assay for the rapid diagnosis of neonatal late onset sepsis. J. Microbiol. Methods 2014, 106, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.R.; Colson, J.D.; Rhoads, D.D. Recent advances in rapid antimicrobial susceptibility testing systems. Expert Rev. Mol. Diagn. 2021, 21, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Sundsfjord, A.; Simonsen, G.S.; Haldorsen, B.C.; Haaheim, H.; Hjelmevoll, S.O.; Littauer, P.; Dahl, K.H. Genetic methods for detection of antimicrobial resistance. Apmis 2004, 112, 815–837. [Google Scholar] [CrossRef]
- Wada, K.; Mizoguchi, S.; Ito, Y.; Kawada, J.; Yamauchi, Y.; Morishima, T.; Nishiyama, Y.; Kimura, H. Multiplex real-time PCR for the simultaneous detection of herpes simplex virus, human herpesvirus 6, and human herpesvirus 7. Microbiol. Immunol. 2009, 53, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Theodore, M.J.; Mair, R.; Trujillo-Lopez, E.; du Plessis, M.; Wolter, N.; Baughman, A.L.; Hatcher, C.; Vuong, J.; Lott, L.; et al. Clinical Validation of Multiplex Real-Time PCR Assays for Detection of Bacterial Meningitis Pathogens. J. Clin. Microbiol. 2012, 50, 702–708. [Google Scholar] [CrossRef]
- Trung, N.T.; Tong, H.V.; Lien, T.T.; Son, T.V.; Huyen, T.T.T.; Quyen, D.T.; Hoan, P.Q.; Meyer, C.G.; Song, L.H. Clinical utility of an optimised multiplex real-time PCR assay for the identification of pathogens causing sepsis in Vietnamese patients. Int. J. Infect. Dis. 2018, 67, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.S.; McClure, J.T.; Mangold, K.; Peterson, L.R. Performance of 3 real-time PCR assays for direct detection of Staphylococcus aureus and MRSA from clinical samples. Diagn. Microbiol. Infect. Dis. 2015, 83, 211–215. [Google Scholar] [CrossRef]
- Rossney, A.S.; Herra, C.M.; Brennan, G.I.; Morgan, P.M.; O’Connell, B. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J. Clin. Microbiol. 2008, 46, 3285–3290. [Google Scholar] [CrossRef]
- Jung, M.K.; Lee, W.G.; Park, M.H. Evaluation of a newly developed multiplex real-time PCR assay for the detection of vancomycin-resistant enterococci from rectal swabs. Korean J. Clin. Microbiol. 2011, 14, 138–147. [Google Scholar] [CrossRef][Green Version]
- Bae, M.H.; Kim, J.; Sung, H.; Jeong, Y.S.; Kim, M.N. Evaluation of iNtRON VRE vanA/vanB real-time PCR for follow-up surveillance of VRE-infected or colonized patient. Diagn. Microbiol. Infect. Dis. 2013, 77, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Chiba, M.; Aoyagi, T.; Yoshida, M.; Katsumi, M.; Fujimaki, S.I.; Ishii, Y.; Tateda, K.; Kaku, M. Evaluation of the performance of GeneSoC®, a novel rapid real-time PCR system, to detect Staphylococcus aureus and methicillin resistance in blood cultures. Infect. Chemother. 2023, 29, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Westh, H.; Lisby, G.; Breysse, F.; Böddinghaus, B.; Chomarat, M.; Gant, V.; Goglio, A.; Raglio, A.; Schuster, H.; Stuber, F.; et al. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin. Microbiol. Infect. 2009, 15, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Samuel, L. Direct Detection of Pathogens in Bloodstream During Sepsis: Are We There Yet? J. Appl. Lab. Med. 2019, 3, 631–642. [Google Scholar] [CrossRef] [PubMed]
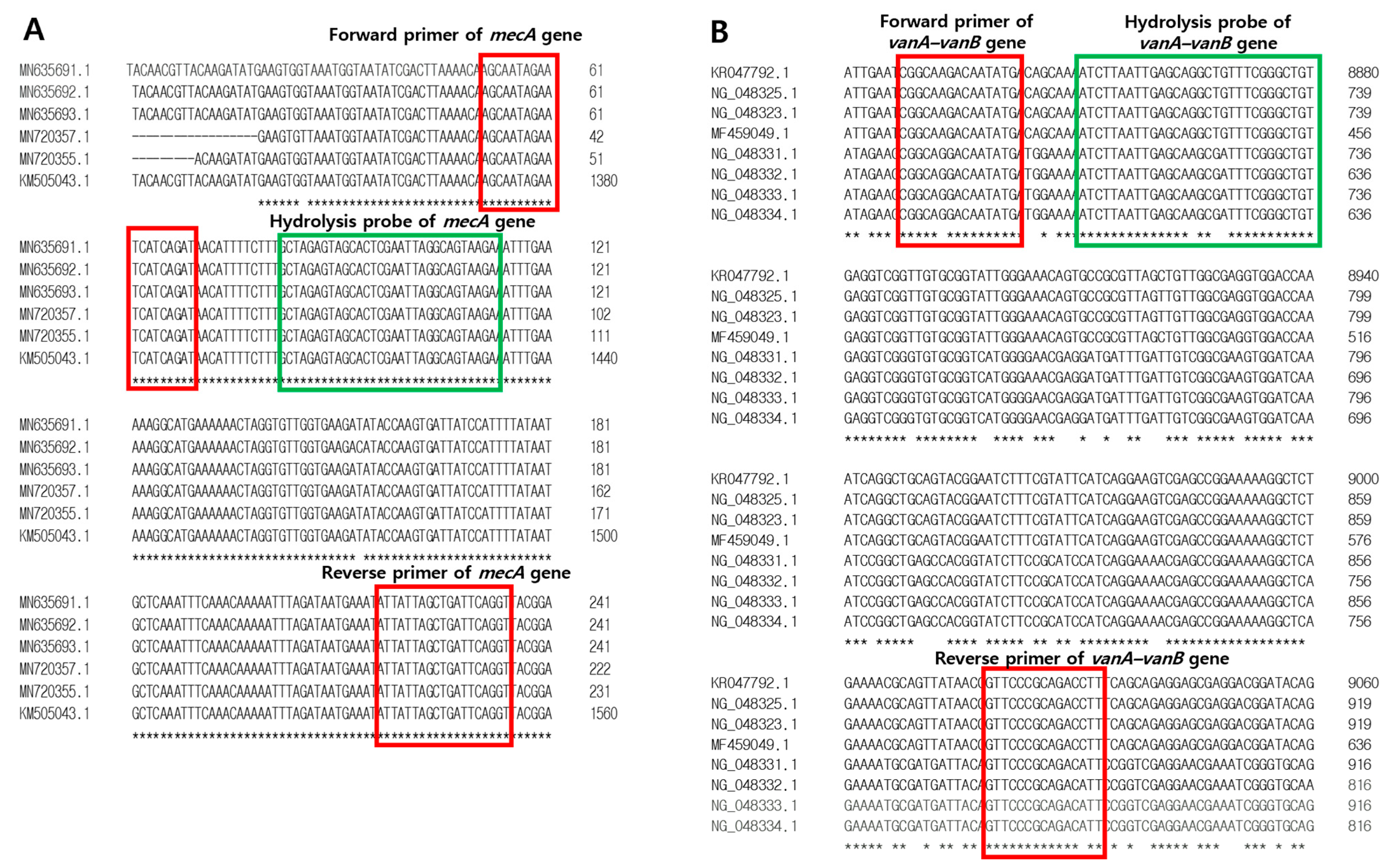



| Target | Primers and Probes | Sequences (5′-3′) | Amplicon Size (bp) | Length (mer) |
|---|---|---|---|---|
| mecA | Forward primer | AGCAATAGAATCATCAGAT | 184 | 19 |
| Reverse primer | ACCTGAATCAGCTAATAAT | 19 | ||
| Probe | FAM-GCTAGAGTAGCACTCGAATTAGGCAGTAAGA-TAMRA | 31 | ||
| vanA-vanB | Forward primer | CGGCAGGACAATATGA | 206 | 16 |
| Reverse primer | AATGTCTGCGGGAAC | 15 | ||
| Probe | HEX-ATCTTAATTGAGCAAGCGATTTCGGGCTGT-TAMRA | 30 |
| Antimicrobial Agent | Standard Interpretation (CLSI) [14] | Total n (%) | ||
|---|---|---|---|---|
| Resistant n (%) | Intermediate n (%) | Susceptible n (%) | ||
| Oxacillin | 52 (30.2%) | 0 (0%) | 39 (22.7%) | 172 (100%) |
| Vancomycin | 80 (46.5%) | 0 (0%) | 1 (0.6%) | |
| Plasmid DNA of Resistance Gene | Plasmid DNA Content | Mean Ct Value | SD | Log Value | Copies per Reaction (copies/μL) |
|---|---|---|---|---|---|
| mecA gene | 2.58 ng | 9.04 | 0.00 | 8.02 | 1.04 × 108 |
| 258 pg | 12.18 | 0.18 | 7.11 | 1.28 × 107 | |
| 25.8 pg | 16.09 | 0.11 | 5.97 | 9.42 × 105 | |
| 2.58 pg | 19.53 | 0.17 | 4.98 | 9.49 × 104 | |
| 258 fg | 23.15 | 0.18 | 3.93 | 8.48 × 103 | |
| 25.8 fg | 26.70 | 0.00 | 2.90 | 7.94 × 102 | |
| 2.58 fg | 30.17 | 0.09 | 1.89 | 7.84 × 101 | |
| 258 ag | 32.54 | 0.32 | 1.21 | 1.61 × 101 | |
| vanA-vanB gene | 964 pg | 11.61 | 0.11 | 8.03 | 1.07 × 108 |
| 96.4 pg | 15.15 | 0.07 | 6.96 | 9.22 × 106 | |
| 9.64 pg | 18.26 | 0.07 | 6.03 | 1.07 × 106 | |
| 964 fg | 21.72 | 0.14 | 4.99 | 9.66 × 104 | |
| 96.4 fg | 25.25 | 0.05 | 3.92 | 8.31 × 103 | |
| 9.64 fg | 28.17 | 0.21 | 3.04 | 1.10 × 103 | |
| 964 ag | 31.14 | 0.01 | 2.15 | 1.40 × 102 | |
| 96.4 ag | 35.26 | 0.24 | 0.9 | 0.83 × 100 |
| Clinical Isolates | Mutiplex qPCR Using mecA and vanA-vanB Primers-Hydrolysis Probes | Sequencing Results Using mecA Primer, vanA-vanB Primer, and Universal Primer | |||||
|---|---|---|---|---|---|---|---|
| mecA Positive Ct Mean ± SD (n, %) | mecA Negative Ct Mean ± SD (n, %) | vanA-vanB Positive Ct Mean ± SD (n, %) | vanA-vanB Negative Ct Mean ± SD (n, %) | mecA Positive Using mecA Primer (n, %) | van Positive Using vanA-vanB Primer (n, %) | Using Universal Primer (n, %) | |
| MRSA isolates (n = 52, 100%) | 20.7 ± 2.3 (50, 96.2%) | 40.0 ± 0.0 (2, 3.8%) | (0, 0%) | 40.0 ± 0.0 (52, 100%) | (50, 96.2%) | - | (52, 100%) |
| MSSA isolates (n = 39, 100%) | (0, 0%) | 40.0 ± 0.2 (39, 100%) | (0, 0%) | 40.0 ± 0.0 (39, 100%) | - | - | - |
| VRE isolates (n = 80, 100%) | (0, 0%) | 39.8 ± 1.2 (80, 100%) | 18.3 ± 1.4 (80, 100%) | (0, 0%) | - | (80, 100%) | (80, 100%) |
| VSE isolate (n = 1, 100%) | (0, 0%) | 40.0 ± 0.0 (1, 100%) | 16.8 ± 0.0 (1, 100%) | (0, 0%) | - | (1, 100%) | (1, 100%) |
| Total isolates (n = 172, 100%) | 20.7 ± 2.3 (50, 29.1%) | 39.9 ± 1.0 (122, 70.9%) | 18.3 ± 1.4 (81, 47.1%) | 40.0 ± 0.0 (91, 52.9%) | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Baek, E.; Ahn, H.; Bae, J.; Kim, S.; Kim, S.; Lee, S.; Kim, S. Development of a One-Step Multiplex qPCR Assay for Detection of Methicillin and Vancomycin Drug Resistance Genes in Antibiotic-Resistant Bacteria. Pathogens 2024, 13, 853. https://doi.org/10.3390/pathogens13100853
Lee J, Baek E, Ahn H, Bae J, Kim S, Kim S, Lee S, Kim S. Development of a One-Step Multiplex qPCR Assay for Detection of Methicillin and Vancomycin Drug Resistance Genes in Antibiotic-Resistant Bacteria. Pathogens. 2024; 13(10):853. https://doi.org/10.3390/pathogens13100853
Chicago/Turabian StyleLee, Jiyoung, Eunyoung Baek, Hyesun Ahn, Jinyoung Bae, Sangha Kim, Sohyeong Kim, Suchan Lee, and Sunghyun Kim. 2024. "Development of a One-Step Multiplex qPCR Assay for Detection of Methicillin and Vancomycin Drug Resistance Genes in Antibiotic-Resistant Bacteria" Pathogens 13, no. 10: 853. https://doi.org/10.3390/pathogens13100853
APA StyleLee, J., Baek, E., Ahn, H., Bae, J., Kim, S., Kim, S., Lee, S., & Kim, S. (2024). Development of a One-Step Multiplex qPCR Assay for Detection of Methicillin and Vancomycin Drug Resistance Genes in Antibiotic-Resistant Bacteria. Pathogens, 13(10), 853. https://doi.org/10.3390/pathogens13100853






