H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host
Abstract
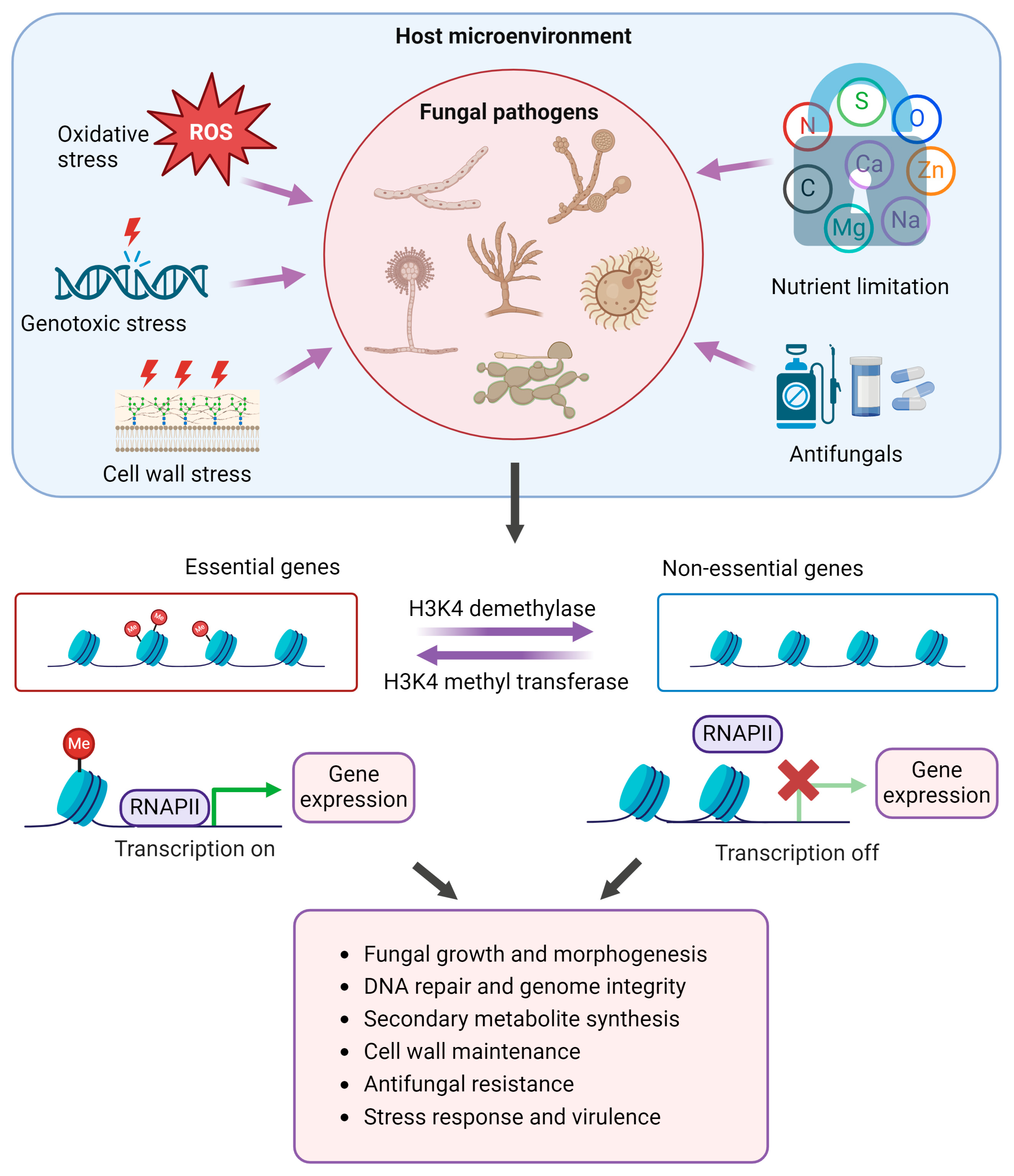
:1. Introduction
1.1. Histone Modifications and Chromatin Architecture
1.2. Histone Methylation
1.3. H3K4 Methylation and Demethylation
1.4. Roles of H3K4 Methylation and Demethylation in Pathogenic Adaptation in Fungi
- (a)
- Morphogenesis and development
- (b)
- Genome stability and DNA repair
- (c)
- Metabolic adaptation
- (d)
- Cell wall maintenance
- (e)
- Antifungal resistance
- (f)
- Stress response and virulence
2. Discussion and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Janbon, G.; Quintin, J.; Lanternier, F.; d’Enfert, C. Studying Fungal Pathogens of Humans and Fungal Infections: Fungal Diversity and Diversity of Approaches. Genes Immun. 2019, 20, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A. Evolution of the Human Pathogenic Lifestyle in Fungi. Nat. Microbiol. 2022, 7, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Benedict, K.; Richardson, M.; Vallabhaneni, S.; Jackson, B.R.; Chiller, T. Emerging Issues, Challenges, and Changing Epidemiology of Fungal Disease Outbreaks. Lancet Infect. Dis. 2017, 17, e403–e411. [Google Scholar] [CrossRef] [PubMed]
- Avery, S.V.; Singleton, I.; Magan, N.; Goldman, G.H. The Fungal Threat to Global Food Security. Fungal Biol. 2019, 123, 555–557. [Google Scholar] [CrossRef]
- Davies, C.R.; Wohlgemuth, F.; Young, T.; Violet, J.; Dickinson, M.; Sanders, J.-W.; Vallieres, C.; Avery, S.V. Evolving Challenges and Strategies for Fungal Control in the Food Supply Chain. Fungal Biol. Rev. 2021, 36, 15–26. [Google Scholar] [CrossRef]
- Xu, K.; Li, X.-Q.; Zhao, D.-L.; Zhang, P. Antifungal Secondary Metabolites Produced by the Fungal Endophytes: Chemical Diversity and Potential Use in the Development of Biopesticides. Front. Microbiol. 2021, 12, 689527. [Google Scholar] [CrossRef]
- Singh, Y.; Nair, A.M.; Verma, P.K. Surviving the Odds: From Perception to Survival of Fungal Phytopathogens under Host-Generated Oxidative Burst. Plant Commun. 2021, 2, 100142. [Google Scholar] [CrossRef]
- Brown, G.D. Innate Antifungal Immunity: The Key Role of Phagocytes. Annu. Rev. Immunol. 2011, 29, 1–21. [Google Scholar] [CrossRef]
- Romani, L. Immunity to Fungal Infections. Nat. Rev. Immunol. 2011, 11, 275–288. [Google Scholar] [CrossRef]
- Sethiya, P.; Rai, M.N.; Rai, R.; Parsania, C.; Tan, K.; Wong, K.H. Transcriptomic Analysis Reveals Global and Temporal Transcription Changes during Candida glabrata Adaptation to an Oxidative Environment. Fungal Biol. 2020, 124, 427–439. [Google Scholar] [CrossRef]
- Rai, M.N.; Balusu, S.; Gorityala, N.; Dandu, L.; Kaur, R. Functional Genomic Analysis of Candida glabrata-Macrophage Interaction: Role of Chromatin Remodeling in Virulence. PLoS Pathog. 2012, 8, e1002863. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S. To Be or Not To Be Pathogenic: Transcriptional Reprogramming Dictates a Fungal Pathogen’s Response to Different Hosts[OPEN]. Plant Cell 2020, 32, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Jenull, S.; Tscherner, M.; Mair, T.; Kuchler, K. ATAC-Seq Identifies Chromatin Landscapes Linked to the Regulation of Oxidative Stress in the Human Fungal Pathogen Candida albicans. J. Fungi 2020, 6, 182. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; Joosten, L.A.B.; van der Meer, J.W.M.; Kullberg, B.-J.; van de Veerdonk, F.L. Immune Defence against Candida Fungal Infections. Nat. Rev. Immunol. 2015, 15, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Quintin, J.; Saeed, S.; Martens, J.H.A.; Giamarellos-Bourboulis, E.J.; Ifrim, D.C.; Logie, C.; Jacobs, L.; Jansen, T.; Kullberg, B.-J.; Wijmenga, C.; et al. Candida Albicans Infection Affords Protection against Reinfection via Functional Reprogramming of Monocytes. Cell Host Microbe 2012, 12, 223–232. [Google Scholar] [CrossRef]
- Dubey, A.; Jeon, J. Epigenetic Regulation of Development and Pathogenesis in Fungal Plant Pathogens. Mol. Plant Pathol. 2017, 18, 887–898. [Google Scholar] [CrossRef]
- Rando, O.J.; Winston, F. Chromatin and Transcription in Yeast. Genetics 2012, 190, 351–387. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Kouzarides, T. Chromatin Modifications and Their Function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Millán-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone Post-Translational Modifications—Cause and Consequence of Genome Function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef] [PubMed]
- Weiner, A.; Chen, H.V.; Liu, C.L.; Rahat, A.; Klien, A.; Soares, L.; Gudipati, M.; Pfeffner, J.; Regev, A.; Buratowski, S.; et al. Systematic Dissection of Roles for Chromatin Regulators in a Yeast Stress Response. PLoS Biol. 2012, 10, e1001369. [Google Scholar] [CrossRef] [PubMed]
- South, P.F.; Harmeyer, K.M.; Serratore, N.D.; Briggs, S.D. H3K4 Methyltransferase Set1 Is Involved in Maintenance of Ergosterol Homeostasis and Resistance to Brefeldin A. Proc. Natl. Acad. Sci. USA 2013, 110, E1016–E1025. [Google Scholar] [CrossRef]
- Lai, Y.; Cao, X.; Chen, J.; Wang, L.; Wei, G.; Wang, S. Coordinated Regulation of Infection-Related Morphogenesis by the KMT2-Cre1-Hyd4 Regulatory Pathway to Facilitate Fungal Infection. Sci. Adv. 2023, 6, eaaz1659. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, M.; Xie, R.; Zhang, F.; Wang, S.; Pan, X.; Wang, S.; Zhuang, Z. The Methyltransferase AflSet1 Is Involved in Fungal Morphogenesis, AFB1 Biosynthesis, and Virulence of Aspergillus Flavus. Front. Microbiol. 2020, 11, 234. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, X.; Sun, W.; Zhang, M.; Yin, Y.; Pan, S.; He, D.; Shen, M.; Yang, J.; Zheng, Q.; et al. The COMPASS-like Complex Modulates Fungal Development and Pathogenesis by Regulating H3K4me3-Mediated Targeted Gene Expression in Magnaporthe oryzae. Mol. Plant Pathol. 2021, 22, 422–439. [Google Scholar] [CrossRef]
- Pehrson, J.R.; Fuji, R.N. Evolutionary Conservation of Histone MacroH2A Subtypes and Domains. Nucleic Acids Res. 1998, 26, 2837–2842. [Google Scholar] [CrossRef]
- Phillips, E.O.N.; Gunjan, A. Histone Variants: The Unsung Guardians of the Genome. DNA Repair 2022, 112, 103301. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Kuo, M.-H.; Allis, C.D. Roles of Histone Acetyltransferases and Deacetylases in Gene Regulation. BioEssays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Kurdistani, S.K.; Grunstein, M. Histone Acetylation and Deacetylation in Yeast. Nat. Rev. Mol. Cell Biol. 2003, 4, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone Lysine Methylation Dynamics: Establishment, Regulation, and Biological Impact. Mol. Cell 2012, 48, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Separovich, R.J.; Wilkins, M.R. Ready, SET, Go: Post-Translational Regulation of the Histone Lysine Methylation Network in Budding Yeast. J. Biol. Chem. 2021, 297, 100939. [Google Scholar] [CrossRef] [PubMed]
- Separovich, R.J.; Pang, C.N.I.; Wilkins, M.R. Controlling the Controllers: Regulation of Histone Methylation by Phosphosignalling. Trends Biochem. Sci. 2020, 45, 1035–1048. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Colabardini, A.C.; Wang, F.; Miao, Z.; Pardeshi, L.; Valero, C.; de Castro, P.A.; Akiyama, D.Y.; Tan, K.; Nora, L.C.; Silva-Rocha, R.; et al. Chromatin Profiling Reveals Heterogeneity in Clinical Isolates of the Human Pathogen Aspergillus fumigatus. PLoS Genet. 2022, 18, e1010001. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Humphrey, E.L.; Erlich, R.L.; Schneider, R.; Bouman, P.; Liu, J.S.; Kouzarides, T.; Schreiber, S.L. Methylation of Histone H3 Lys 4 in Coding Regions of Active Genes. Proc. Natl. Acad. Sci. USA 2002, 99, 8695–8700. [Google Scholar] [CrossRef]
- Kouzarides, T. Histone Methylation in Transcriptional Control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [Google Scholar] [CrossRef]
- Howe, F.S.; Fischl, H.; Murray, S.C.; Mellor, J. Is H3K4me3 Instructive for Transcription Activation? Bioessays 2017, 39, 1–12. [Google Scholar] [CrossRef]
- Ng, H.H.; Robert, F.; Young, R.A.; Struhl, K. Targeted Recruitment of Set1 Histone Methylase by Elongating Pol II Provides a Localized Mark and Memory of Recent Transcriptional Activity. Mol. Cell 2003, 11, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.-W.; Allis, C.D. Ubiquitination of Histone H2B Regulates H3 Methylation and Gene Silencing in Yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Dover, J.; Schneider, J.; Tawiah-Boateng, M.A.; Wood, A.; Dean, K.; Johnston, M.; Shilatifard, A. Methylation of Histone H3 by COMPASS Requires Ubiquitination of Histone H2B by Rad6. J. Biol. Chem. 2002, 277, 28368–28371. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.; Bryk, M. Diverse and Dynamic Forms of Gene Regulation by the S. cerevisiae Histone Methyltransferase Set1. Curr. Genet. 2023, 69, 91–114. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Whetstine, J.R. Dynamic Regulation of Histone Lysine Methylation by Demethylases. Mol. Cell 2007, 25, 1–14. [Google Scholar] [CrossRef]
- Liang, G.; Klose, R.J.; Gardner, K.E.; Zhang, Y. Yeast Jhd2p Is a Histone H3 Lys4 Trimethyl Demethylase. Nat. Struct. Mol. Biol. 2007, 14, 243–245. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Pokhrel, S.; Palani, S.; Pflueger, C.; Parnell, T.J.; Cairns, B.R.; Bhaskara, S.; Chandrasekharan, M.B. Counteracting H3K4 Methylation Modulators Set1 and Jhd2 Co-Regulate Chromatin Dynamics and Gene Transcription. Nat. Commun. 2016, 7, 11949. [Google Scholar] [CrossRef]
- Dallery, J.-F.; Adelin, É.; Le Goff, G.; Pigné, S.; Auger, A.; Ouazzani, J.; O’Connell, R.J. H3K4 Trimethylation by CclA Regulates Pathogenicity and the Production of Three Families of Terpenoid Secondary Metabolites in Colletotrichum higginsianum. Mol. Plant Pathol. 2019, 20, 831–842. [Google Scholar] [CrossRef]
- Liu, R.; Chen, X.; Zhao, F.; Jiang, Y.; Lu, Z.; Ji, H.; Feng, Y.; Li, J.; Zhang, H.; Zheng, J.; et al. The COMPASS Complex Regulates Fungal Development and Virulence through Histone Crosstalk in the Fungal Pathogen Cryptococcus neoformans. J. Fungi 2023, 9, 672. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, N.; Yin, Y.; Chen, Y.; Jiang, J.; Ma, Z. Histone H3K4 Methylation Regulates Hyphal Growth, Secondary Metabolism and Multiple Stress Responses in Fusarium graminearum. Environ. Microbiol. 2015, 17, 4615–4630. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Li, B.; Tian, S. Histone H3K4 Methyltransferase PeSet1 Regulates Colonization, Patulin Biosynthesis, and Stress Responses of Penicillium expansum. Microbiol. Spectr. 2023, 11, e03545-22. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Fang, W.; Tong, J.; Liu, S.; Wu, H.; Shi, J. Metarhizium robertsii as a Promising Microbial Agent for Rice in Situ Cadmium Reduction and Plant Growth Promotion. Chemosphere 2022, 305, 135427. [Google Scholar] [CrossRef]
- Meng, S.; Shi, H.; Lin, C.; Wu, Z.; Lin, F.; Tao, Z.; Kou, Y. UvKmt2-Mediated H3K4 Trimethylation Is Required for Pathogenicity and Stress Response in Ustilaginoidea virens. J. Fungi 2022, 8, 553. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.-W.; Ahn, S.H. Role of Yeast JmjC-Domain Containing Histone Demethylases in Actively Transcribed Regions. Biochem. Biophys. Res. Commun. 2011, 410, 614–619. [Google Scholar] [CrossRef] [PubMed]
- Faucher, D.; Wellinger, R.J. Methylated H3K4, a Transcription-Associated Histone Modification, Is Involved in the DNA Damage Response Pathway. PLoS Genet. 2010, 6, e1001082. [Google Scholar] [CrossRef] [PubMed]
- Chong, S.Y.; Cutler, S.; Lin, J.-J.; Tsai, C.-H.; Tsai, H.-K.; Biggins, S.; Tsukiyama, T.; Lo, Y.-C.; Kao, C.-F. H3K4 Methylation at Active Genes Mitigates Transcription-Replication Conflicts during Replication Stress. Nat. Commun. 2020, 11, 809. [Google Scholar] [CrossRef]
- Choudhary, N.K.; Jain, A.K.; Soni, R.; Gahlot, N. Mucormycosis: A Deadly Black Fungus Infection among COVID-19 Patients in India. Clin. Epidemiol. Glob. Health 2021, 12, 100900. [Google Scholar] [CrossRef]
- Osorio-Concepción, M.; Lax, C.; Lorenzo-Gutiérrez, D.; Cánovas-Márquez, J.T.; Tahiri, G.; Navarro, E.; Binder, U.; Nicolás, F.E.; Garre, V. H3K4 Methylation Regulates Development, DNA Repair, and Virulence in Mucorales. IMA Fungus 2023, 15, 6. [Google Scholar] [CrossRef]
- Ren, K.; Mou, Y.-N.; Tong, S.-M.; Ying, S.-H.; Feng, M.-G. DIM5/KMT1 Controls Fungal Insect Pathogenicity and Genome Stability by Methylation of Histone H3K4, H3K9 and H3K36. Virulence 2021, 12, 1306–1322. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Brakhage, A.A. Human and Plant Fungal Pathogens: The Role of Secondary Metabolites. PLoS Pathog. 2014, 10, e1003859. [Google Scholar] [CrossRef]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Bachleitner, S.; Sørensen, J.L.; Gacek-Matthews, A.; Sulyok, M.; Studt, L.; Strauss, J. Evidence of a Demethylase-Independent Role for the H3K4-Specific Histone Demethylases in Aspergillus nidulans and Fusarium graminearum Secondary Metabolism. Front. Microbiol. 2019, 10, 1759. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Bok, J.W.; Lee, S.; Dagenais, T.R.T.; Andes, D.R.; Kontoyiannis, D.P.; Keller, N.P. Loss of CclA, Required for Histone 3 Lysine 4 Methylation, Decreases Growth but Increases Secondary Metabolite Production in Aspergillus fumigatus. PeerJ 2013, 1, e4. [Google Scholar] [CrossRef] [PubMed]
- Gacek-Matthews, A.; Berger, H.; Sasaki, T.; Wittstein, K.; Gruber, C.; Lewis, Z.A.; Strauss, J. KdmB, a Jumonji Histone H3 Demethylase, Regulates Genome-Wide H3K4 Trimethylation and Is Required for Normal Induction of Secondary Metabolism in Aspergillus nidulans. PLoS Genet. 2016, 12, e1006222. [Google Scholar] [CrossRef]
- Baker, K.M.; Hoda, S.; Saha, D.; Gregor, J.B.; Georgescu, L.; Serratore, N.D.; Zhang, Y.; Cheng, L.; Lanman, N.A.; Briggs, S.D. The Set1 Histone H3K4 Methyltransferase Contributes to Azole Susceptibility in a Species-Specific Manner by Differentially Altering the Expression of Drug Efflux Pumps and the Ergosterol Gene Pathway. Antimicrob. Agents Chemother. 2022, 66, e0225021. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.; Kwon, S.; Lee, E.-J.; Lee, J.-S. Set1-Mediated H3K4 Methylation Is Required for Candida albicans Virulence by Regulating Intracellular Level of Reactive Oxygen Species. Virulence 2021, 12, 2648–2658. [Google Scholar] [CrossRef]
- Gacek-Matthews, A.; Noble, L.M.; Gruber, C.; Berger, H.; Sulyok, M.; Marcos, A.T.; Strauss, J.; Andrianopoulos, A. KdmA, a Histone H3 Demethylase with Bipartite Function, Differentially Regulates Primary and Secondary Metabolism in Aspergillus nidulans. Mol. Microbiol. 2015, 96, 839–860. [Google Scholar] [CrossRef]
- Choi, Y.-H.; Lee, M.-W.; Shin, K.-S. The Lysine Demethylases KdmA and KdmB Differently Regulate Asexual Development, Stress Response, and Virulence in Aspergillus fumigatus. J. Fungi 2022, 8, 590. [Google Scholar] [CrossRef]
- Meng, S.; Huang, S.; Liu, J.; Gai, Y.; Li, M.; Duan, S.; Zhang, S.; Sun, X.; Yang, Q.; Wang, Y.; et al. Histone Methylation Is Required for Virulence, Conidiation, and Multi-Stress Resistance of Alternaria alternata. Front. Microbiol. 2022, 13, 924476. [Google Scholar] [CrossRef]
- Hou, J.; Feng, H.-Q.; Chang, H.-W.; Liu, Y.; Li, G.-H.; Yang, S.; Sun, C.-H.; Zhang, M.-Z.; Yuan, Y.; Sun, J.; et al. The H3K4 Demethylase Jar1 Orchestrates ROS Production and Expression of Pathogenesis-Related Genes to Facilitate Botrytis cinerea Virulence. New Phytol. 2020, 225, 930–947. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Wang, L.; Zheng, W.; Wang, S. Regulatory Roles of Histone Modifications in Filamentous Fungal Pathogens. J. Fungi 2022, 8, 565. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fan, Z.; Shliaha, P.V.; Miele, M.; Hendrickson, R.C.; Jiang, X.; Helin, K. H3K4me3 Regulates RNA Polymerase II Promoter-Proximal Pause-Release. Nature 2023, 615, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.A.J.; Shilatifard, A. Reevaluating the Roles of Histone-Modifying Enzymes and Their Associated Chromatin Modifications in Transcriptional Regulation. Nat. Genet. 2020, 52, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Pham, K.T.M.; Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe oryzae) Regulates Global Gene Expression during Infection-Related Morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar]
- Santos-Rosa, H.; Schneider, R.; Bannister, A.J.; Sherriff, J.; Bernstein, B.E.; Emre, N.C.T.; Schreiber, S.L.; Mellor, J.; Kouzarides, T. Active Genes Are Tri-Methylated at K4 of Histone H3. Nature 2002, 419, 407–411. [Google Scholar] [CrossRef]
- Wozniak, G.G.; Strahl, B.D. Hitting the “Mark”: Interpreting Lysine Methylation in the Context of Active Transcription. Biochim. Biophys. Acta 2014, 1839, 1353–1361. [Google Scholar] [CrossRef]
- Yaseen, I.; White, S.A.; Torres-Garcia, S.; Spanos, C.; Lafos, M.; Gaberdiel, E.; Yeboah, R.; El Karoui, M.; Rappsilber, J.; Pidoux, A.L.; et al. Proteasome-Dependent Truncation of the Negative Heterochromatin Regulator Epe1 Mediates Antifungal Resistance. Nat. Struct. Mol. Biol. 2022, 29, 745–758. [Google Scholar] [CrossRef]
- Patra, S.; Raney, M.; Pareek, A.; Kaur, R. Epigenetic Regulation of Antifungal Drug Resistance. J. Fungi 2022, 8, 875. [Google Scholar] [CrossRef]
- Moirangthem, R.; Kumar, K.; Kaur, R. Two Functionally Redundant FK506-Binding Proteins Regulate Multidrug Resistance Gene Expression and Govern Azole Antifungal Resistance. Antimicrob. Agents Chemother. 2021, 65, 10–1128. [Google Scholar] [CrossRef]
- Torres-Garcia, S.; Yaseen, I.; Shukla, M.; Audergon, P.N.C.B.; White, S.A.; Pidoux, A.L.; Allshire, R.C. Epigenetic Gene Silencing by Heterochromatin Primes Fungal Resistance. Nature 2020, 585, 453–458. [Google Scholar] [CrossRef] [PubMed]
- Raman, S.B.; Nguyen, M.H.; Zhang, Z.; Cheng, S.; Jia, H.Y.; Weisner, N.; Iczkowski, K.; Clancy, C.J. Candida albicans SET1 Encodes a Histone 3 Lysine 4 Methyltransferase That Contributes to the Pathogenesis of Invasive Candidiasis. Mol. Microbiol. 2006, 60, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yang, W.; Liu, Y.; Li, W.; Wang, Y.; Liu, N.; Sheng, C. Jumonji Histone Demethylase Inhibitor JIB-04 as a Broad-Spectrum Antifungal Agent. ACS Infect. Dis. 2022, 8, 1316–1323. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rai, M.N.; Rai, R. H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host. Pathogens 2024, 13, 1080. https://doi.org/10.3390/pathogens13121080
Rai MN, Rai R. H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host. Pathogens. 2024; 13(12):1080. https://doi.org/10.3390/pathogens13121080
Chicago/Turabian StyleRai, Maruti Nandan, and Rikky Rai. 2024. "H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host" Pathogens 13, no. 12: 1080. https://doi.org/10.3390/pathogens13121080
APA StyleRai, M. N., & Rai, R. (2024). H3K4 Methylation and Demethylation in Fungal Pathogens: The Epigenetic Toolbox for Survival and Adaptation in the Host. Pathogens, 13(12), 1080. https://doi.org/10.3390/pathogens13121080





