Analysis of Efflux Pump Contributions and Plasmid-Mediated Genetic Determinants in Ciprofloxacin-Resistant Salmonella
Abstract
1. Introduction
2. Materials and Methods
2.1. Ciprofloxacin-Resistant Salmonella Strains
2.2. Effects of Efflux Pump Inhibitor on Ciprofloxacin Resistance
2.3. Plasmid Studies
2.4. Whole-Genome Sequencing and Data Analysis
2.5. Nucleotide Sequence Accession Numbers
3. Results
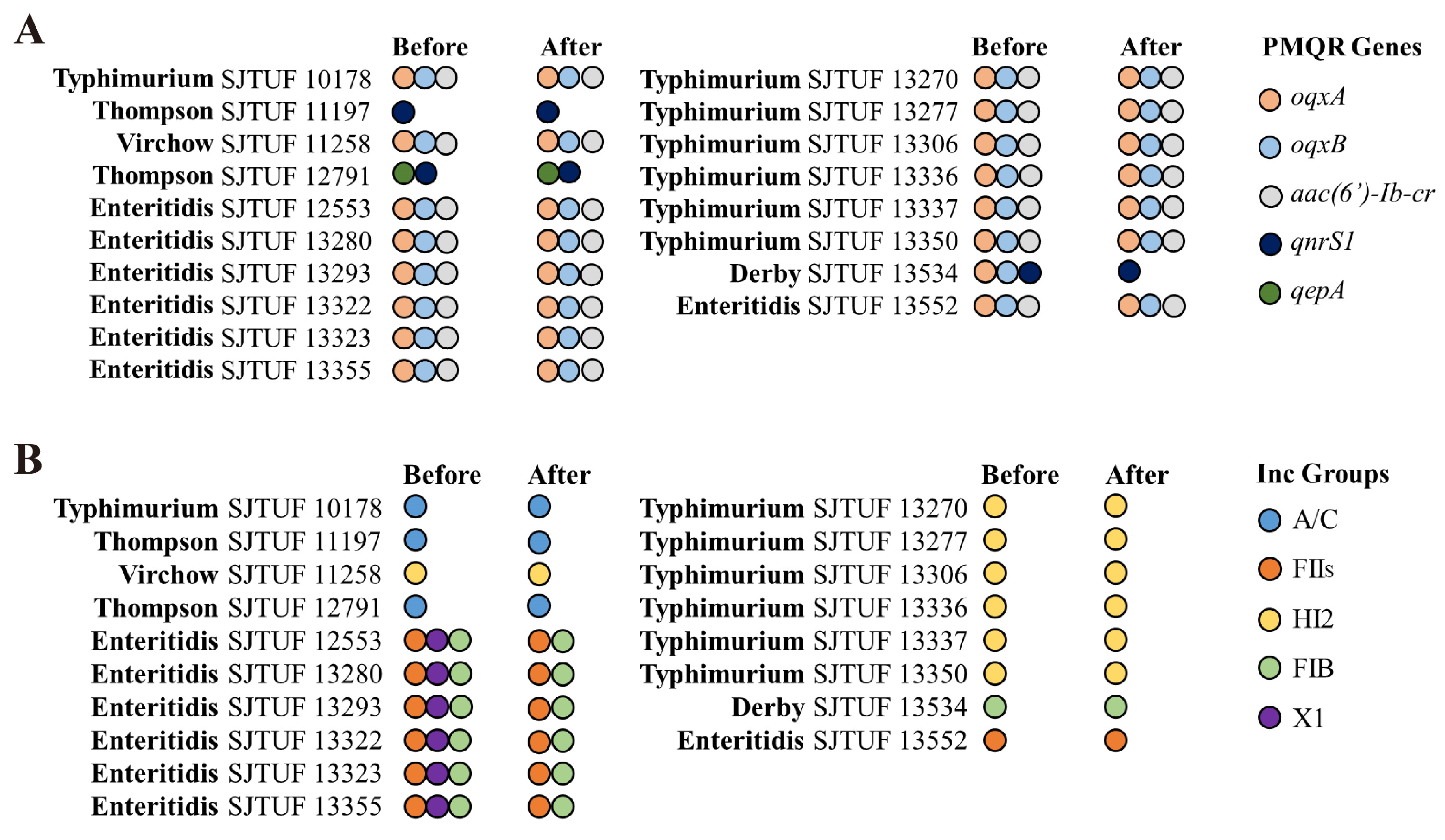
3.1. Correlation Analysis of Molecular Basis for CIPR Salmonella Isolates
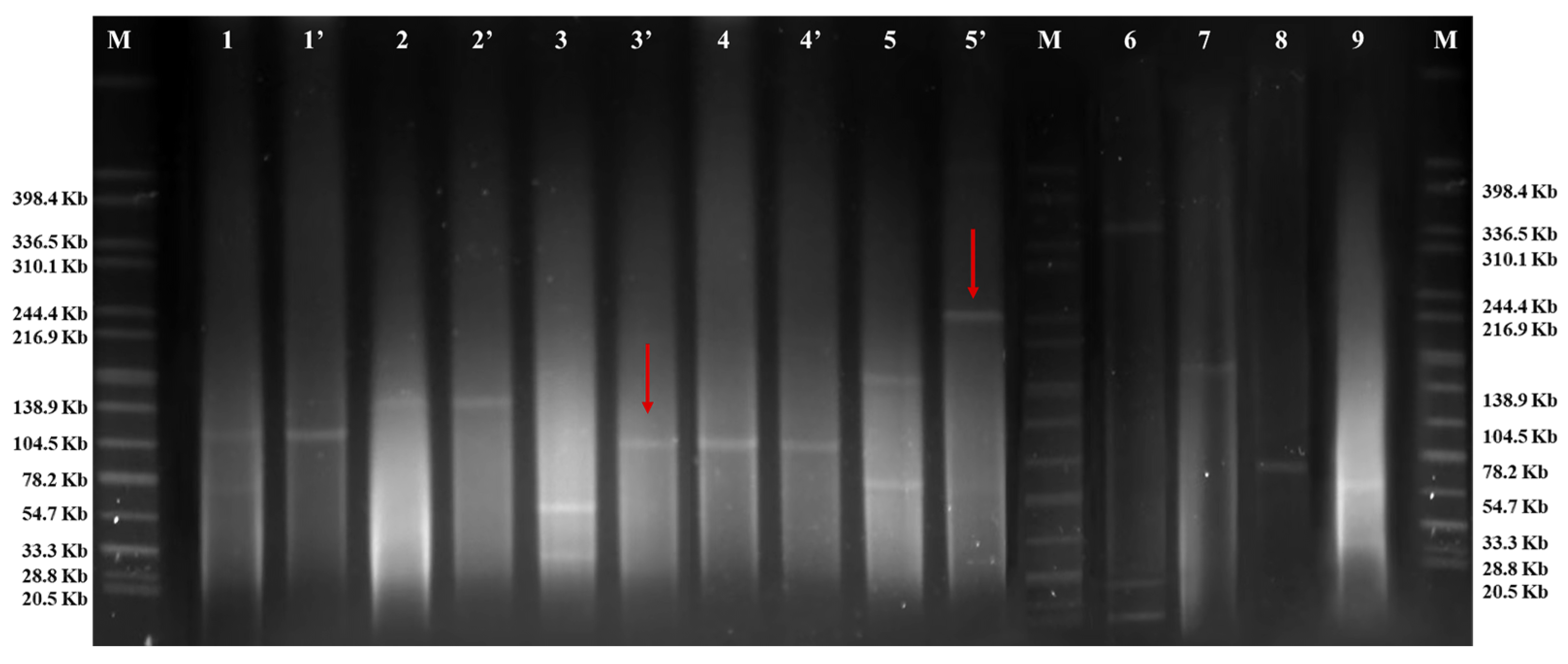
3.2. Transferable Ciprofloxacin-Resistance Genes and Plasmids
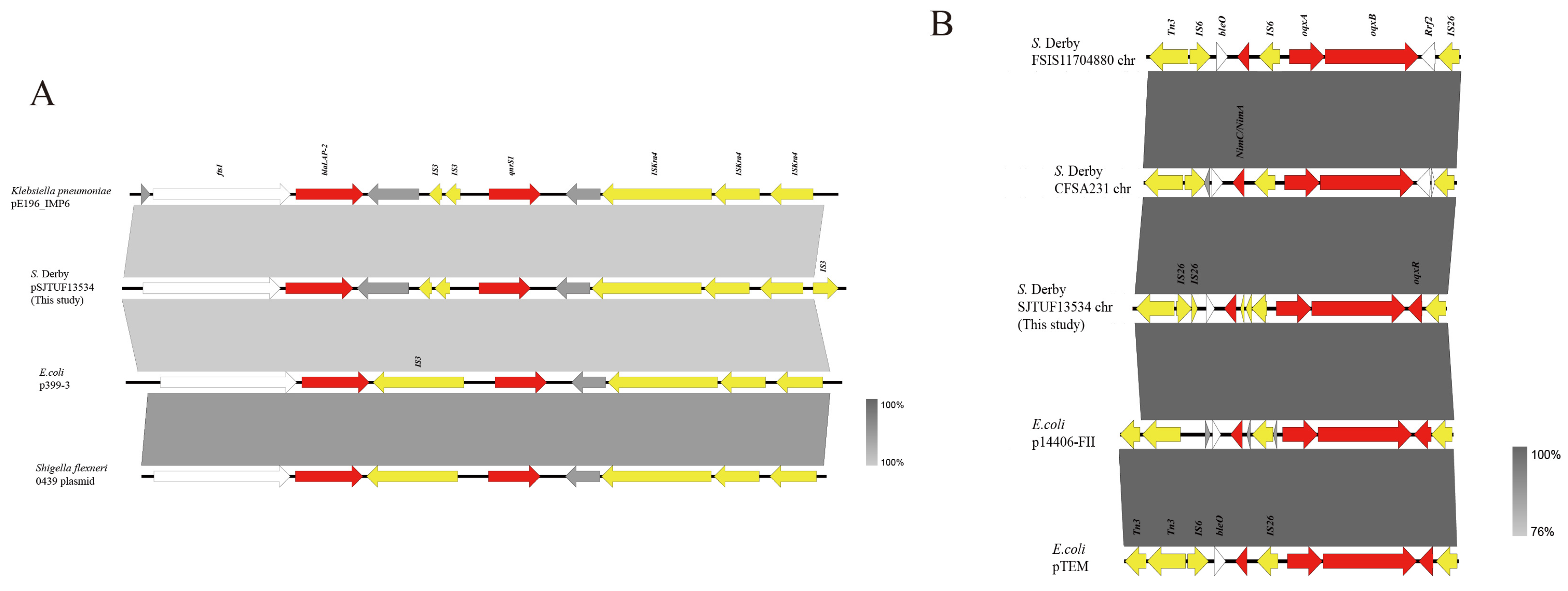
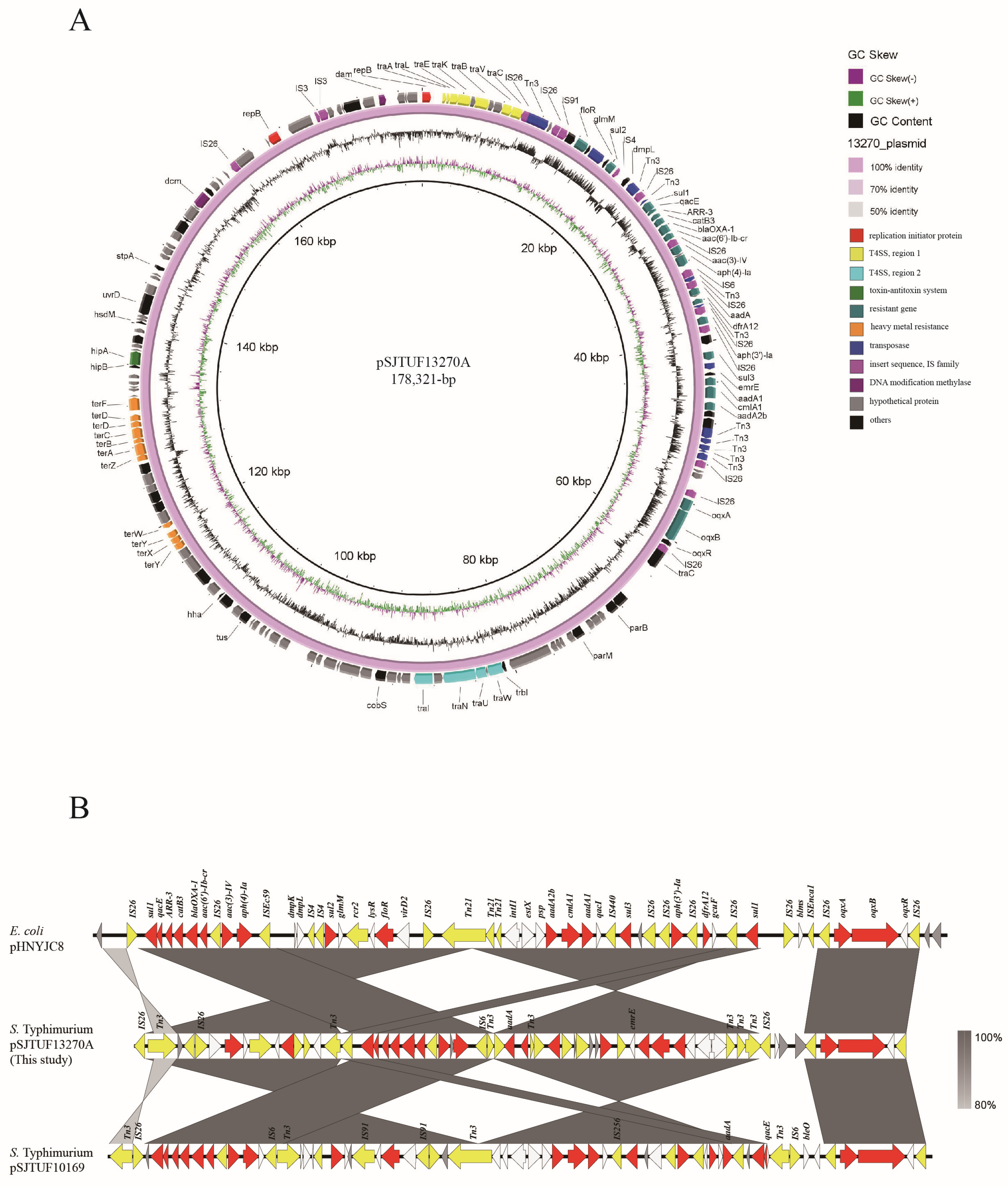
3.3. Complete Sequence of Representative Isolates and Transferable Element Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bai, L.; Li, W.; Han, H.; Fu, P.; Ma, X.; Bi, Z.; Yang, X.; Zhang, X.; Zhen, S.; et al. Trends of foodborne diseases in China: Lessons from laboratory-based surveillance since 2011. Front. Med. 2018, 12, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Shane, A.L.; Mody, R.K.; Crump, J.A.; Tarr, P.I.; Steiner, T.S.; Kotloff, K.; Langley, J.M.; Wanke, C.; Warren, C.A.; Cheng, A.C.; et al. Infectious diseases society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin. Infect. Dis. 2017, 65, e45–e80. [Google Scholar] [CrossRef]
- Ma, Y.; Li, M.; Xu, X.; Fu, Y.; Xiong, Z.; Zhang, L.; Qu, X.; Zhang, H.; Wei, Y.; Zhan, Z.; et al. High-levels of resistance to quinolone and cephalosporin antimicrobials in MDR-ACSSuT Salmonella enterica serovar Enteritidis mainly isolated from patients and foods in Shanghai, China. Int. J. Food Microbiol. 2018, 286, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Z.; Zhou, X.; Cui, Y.; Shi, C.; Shi, X. Prevalence and characterization of antimicrobial resistance in Salmonella enterica isolates from retail foods in Shanghai, China. Foodborne Pathog. Dis. 2020, 17, 35–43. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Mechanisms of drug resistance: Quinolone resistance. Ann. N. Y. Acad. Sci. 2015, 1354, 12–31. [Google Scholar] [CrossRef]
- Horiyama, T.; Yamaguchi, A.; Nishino, K. TolC dependency of multidrug efflux systems in Salmonella enterica serovar Typhimurium. J. Antimicrob. Chemother. 2010, 65, 1372–1376. [Google Scholar] [CrossRef]
- Baucheron, S.; Chaslus-Dancla, E.; Cloeckaert, A. Role of TolC and parC mutation in high-level fluoroquinolone resistance in Salmonella enterica serotype Typhimurium DT204. J. Antimicrob. Chemother. 2004, 53, 657–659. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.M.; Machuca, J.; Cano, M.E.; Calvo, J.; Martínez-Martínez, L.; Pascual, A. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist. Updates 2016, 29, 13–29. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.; Zhang, J.M.; Xu, X.B.; Shi, W.M.; Chen, S.; Yang, X.W.; Su, X.D.; Shi, X.M.; Meng, J.H. Emerging high-level ciprofloxacin resistance and molecular basis of resistance in Salmonella enterica from humans, food and animals. Int. J. Food Microbiol. 2018, 280, 1–9. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Chen, Y.; Hu, D.; Zhang, Q.; Liao, X.P.; Liu, Y.H.; Sun, J. Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell. Infect. Microbiol. 2017, 7, 37. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, Z.L.; Liu, J.H.; Zeng, Z.L.; Ma, J.Y.; Jiang, H.X. Emergence of RmtB methylase producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 2007, 59, 880–885. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Patel, R.; Jain, M. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Li, L.; Liao, X.P.; Yang, Y.R.; Sun, J.; Li, L.L.; Liu, B.T.; Yang, S.S.; Ma, J.; Li, X.; Zhang, Q.J.; et al. Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J. Antimicrob. Chemother. 2013, 68, 2263–2268. [Google Scholar] [CrossRef]
- Huang, L.L.; Wu, C.R.; Gao, H.J.; Xu, C.; Dai, M.H.; Huang, L.L.; Hao, H.H.; Wang, X.; Cheng, G.Y. Bacterial multidrug efflux pumps at the frontline of antimicrobial resistance: An overview. Antibiotics 2022, 11, 520. [Google Scholar] [CrossRef]
- Ballesté-Delpierre, C.; Solé, M.; Domènech, Ò.; Borrell, J.; Vila, J.; Fàbrega, A. Molecular study of quinolone resistance mechanisms and clonal relationship of Salmonella enterica clinical isolates. Int. J. Antimicrob. Agents 2014, 43, 121–125. [Google Scholar] [CrossRef]
- Chen, K.; Dong, N.; Zhao, S.; Liu, L.; Li, R.; Xie, M.; Lin, D.; Wai-Chi Chan, E.; Meng, J.; McDermott, P.F.; et al. Identification and characterization of conjugative plasmids that encode ciprofloxacin resistance in Salmonella. Antimicrob. Agents Chemother. 2018, 62, e00575-18. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.X.; Zhang, J.F.; Sun, Y.H.; Li, R.S.; Lin, X.L.; Yang, L.; Webber, M.A.; Jiang, H.X. Contribution of different mechanisms to ciprofloxacin resistance in Salmonella spp. Front. Microbiol. 2021, 12, 663731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.F.; Yang, J.X.; Xu, X.B.; Zhou, X.J.; Shi, C.L.; Zhao, X.D.; Liu, Y.H.; Shi, X.M. Co-existence of mphA, oqxAB and blaCTX-M-65 on the IncHI2 Plasmid in highly drug-resistant Salmonella enterica serovar Indiana ST17 isolated from retail foods and humans in China. Food Control 2020, 118, 107269. [Google Scholar] [CrossRef]
- Norman, A.; Hansen, L.H.; She, Q.; Sørensen, S.J. Nucleotide sequence of pOLA52: A conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 2008, 60, 59–74. [Google Scholar] [CrossRef]
- Liu, B.T.; Yang, Q.E.; Li, L.; Sun, J.; Liao, X.P.; Fang, L.X.; Yang, S.S.; Deng, H.; Liu, Y.H. Dissemination and characterization of plasmids carrying oqxAB-blaCTX-M genes in Escherichia coli isolates from food-producing animals. PLoS ONE 2013, 8, e73947. [Google Scholar] [CrossRef]
- Shi, H.; Zhou, X.; Zou, W.; Wang, Y.; Lei, C.; Xiang, R.; Zhou, L.; Liu, B.; Zhang, A.; Wang, H. Co-occurrence of biofilm formation and quinolone resistance in Salmonella enterica serotype Typhimurium carrying an IncHI2-type oqxAB-positive plasmid. Microb. Pathog. 2018, 123, 68–73. [Google Scholar] [CrossRef]
- García-Fernández, A.; Fortini, D.; Veldman, K.; Mevius, D.; Carattoli, A. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J. Antimicrob. Chemother. 2009, 63, 274–281. [Google Scholar] [CrossRef]
- Antunes, P.; Mourão, J.; Machado, J.; Peixe, L. First description of qnrS1-IncN plasmid in a ST11 Salmonella Enteritidis clinical isolate from Portugal. Diagn. Microbiol. Infect. Dis. 2011, 69, 463–465. [Google Scholar] [CrossRef]
- Guo, Y.F.; Zhang, W.H.; Ren, S.Q.; Yang, L.; Lü, D.H.; Zeng, Z.L.; Liu, Y.H.; Jiang, H.X. IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PLoS ONE 2014, 9, e96738. [Google Scholar] [CrossRef]
- Ambrose, S.J.; Harmer, C.J.; Hall, R.M. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid 2018, 99, 40–55. [Google Scholar] [CrossRef]
- Chen, K.; Yang, C.; Chan, E.W.; Chen, S. Emergence of conjugative IncC type plasmid simultaneously encoding resistance to ciprofloxacin, ceftriaxone, and azithromycin in Salmonella. Antimicrob. Agents Chemother. 2021, 65, e0104621. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhai, W.S.; Du, P.C.; Wang, Y.; Zhan, Z.F.; Chen, S.; Jia, H.Y.; Bai, L. Molecular characteristics of ciprofloxacin-cefotaxime-azithromycin co-resistant Salmonella enterica serovar Thompson in foodborne diseases in Hunan Province. Zhonghua Yu Fang Yi Xue Za Zhi 2022, 56, 1745–1750. (In Chinese) [Google Scholar] [PubMed]
- Chen, K.; Chan, E.W.C.; Chen, S. Evolution and transmission of a conjugative plasmid encoding both ciprofloxacin and ceftriaxone resistance in Salmonella. Emerg. Microbes Infect. 2019, 8, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Vieira, D.C.; Lima, W.G.; Paiva, M.C. Plasmid-mediated quinolone resistance (PMQR) among Enterobacteriales in Latin America: A systematic review. Mol. Biol. Rep. 2020, 47, 1471–1483. [Google Scholar] [CrossRef]
- Yang, C.; Chen, K.; Chan, E.W.; Yao, W.; Chen, S. Transmission of chromosomal MDR DNA fragment encoding ciprofloxacin resistance by a conjugative helper plasmid in Salmonella. Front. Microbiol. 2020, 11, 556227. [Google Scholar] [CrossRef]
- Chen, K.; Yang, C.; Dong, N.; Xie, M.; Ye, L.; Chan, E.W.C.; Chen, S. Evolution of ciprofloxacin resistance-encoding genetic elements in Salmonella. Msystems 2020, 5, e01234-20. [Google Scholar] [CrossRef]
- Cui, M.Q.; Zhang, P.; Li, J.Y.; Sun, C.T.; Song, L.; Zhang, C.P.; Zhao, Q.; Wu, C.M. Prevalence and characterization of fluoroquinolone resistant Salmonella isolated from an integrated broiler chicken supply chain. Front. Microbiol. 2019, 10, 1865. [Google Scholar] [CrossRef]
- Chen, C.L.; Su, L.H.; Janapatla, R.P.; Lin, C.Y.; Chiu, C.H. Genetic analysis of virulence and antimicrobial-resistant plasmid pOU7519 in Salmonella enterica serovar Choleraesuis. J. Microbiol. Immunol. Infect. 2020, 53, 49–59. [Google Scholar] [CrossRef]
- Gu, Y.X.; Lü, Z.X.; Cao, C.Y.; Sheng, H.J.; Li, W.; Cui, S.H.; Li, R.C.; Lü, X.; Yang, B.W. Cunning plasmid fusion mediates antibiotic resistance genes represented by ESBLs encoding genes transfer in foodborne Salmonella. Int. J. Food Microbiol. 2021, 355, 109336. [Google Scholar] [CrossRef]
| Strains | Serovars | QRDR | Nalidixic Acid | a MIC of Ciprofloxacin (μg/mL) | Fold Changes * | |||
|---|---|---|---|---|---|---|---|---|
| Untreated Salmonella | Treatment with Inhibitor | b E. coli Transconjugants/Transformants | Inhibition | Transconjugant/Transformant | ||||
| SJTUF 13270 | Typhimurium | GyrA (Asp87Asn) | R | 16 | 2 | 4 | 8 | 1/4 |
| SJTUF 13306 | Typhimurium | GyrA (Asp87Asn) | R | 16 | 2 | 4 | 8 | 1/4 |
| SJTUF 13336 | Typhimurium | GyrA (Asp87Asn) | R | 8 | 1 | 2 | 8 | 1/4 |
| SJTUF 13337 | Typhimurium | GyrA (Asp87Asn) | R | 16 | 2 | 4 | 8 | 1/4 |
| SJTUF 13350 | Typhimurium | GyrA (Asp87Asn) | R | 16 | 2 | 4 | 8 | 1/4 |
| SJTUF 13277 | Typhimurium | GyrA (Asp87Asn) | R | 8 | 1 | 2 | 8 | 1/4 |
| SJTUF 10178 | Typhimurium | NA | S | 2 | 0.5 | 1 | 4 | 1/2 |
| SJTUF 13552 | Enteritidis | NA | S | 1 | 0.03 | 1 | 32 | 1 |
| SJTUF 13323 | Enteritidis | GyrA (Asp87Tyr) | R | 8 | 0.25 | 2 | 32 | 1/4 |
| SJTUF 13293 | Enteritidis | GyrA (Asp87Tyr) | R | 8 | 0.25 | 2 | 32 | 1/4 |
| SJTUF 13280 | Enteritidis | GyrA (Asp87Tyr) | R | 8 | 0.25 | 2 | 32 | 1/4 |
| SJTUF 13322 | Enteritidis | GyrA (Asp87Tyr) | R | 8 | 0.5 | 2 | 16 | 1/4 |
| SJTUF 13355 | Enteritidis | GyrA (Asp87Tyr) | R | 8 | 0.25 | 2 | 32 | 1/4 |
| SJTUF 12553 | Enteritidis | NA | S | 1 | 0.06 | 1 | 16 | 1 |
| SJTUF 12791 | Thompson | ParC (Thr57Ser) | S | 128 | 16 | 2 | 8 | 1/64 |
| SJTUF 11197 | Thompson | ParC (Thr57Ser) | S | 64 | 8 | 2 | 8 | 1/32 |
| SJTUF 11258 | Virchow | NA | S | 2 | 0.125 | 1 | 16 | 1/2 |
| SJTUF 13534 | Derby | NA | S | 2 | 0.25 | 2 | 8 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Yi, S.; Kuang, D.; Shi, C.; Qu, C. Analysis of Efflux Pump Contributions and Plasmid-Mediated Genetic Determinants in Ciprofloxacin-Resistant Salmonella. Pathogens 2024, 13, 1126. https://doi.org/10.3390/pathogens13121126
Zhou X, Yi S, Kuang D, Shi C, Qu C. Analysis of Efflux Pump Contributions and Plasmid-Mediated Genetic Determinants in Ciprofloxacin-Resistant Salmonella. Pathogens. 2024; 13(12):1126. https://doi.org/10.3390/pathogens13121126
Chicago/Turabian StyleZhou, Xiujuan, Shanrong Yi, Dai Kuang, Chunlei Shi, and Chunbo Qu. 2024. "Analysis of Efflux Pump Contributions and Plasmid-Mediated Genetic Determinants in Ciprofloxacin-Resistant Salmonella" Pathogens 13, no. 12: 1126. https://doi.org/10.3390/pathogens13121126
APA StyleZhou, X., Yi, S., Kuang, D., Shi, C., & Qu, C. (2024). Analysis of Efflux Pump Contributions and Plasmid-Mediated Genetic Determinants in Ciprofloxacin-Resistant Salmonella. Pathogens, 13(12), 1126. https://doi.org/10.3390/pathogens13121126







