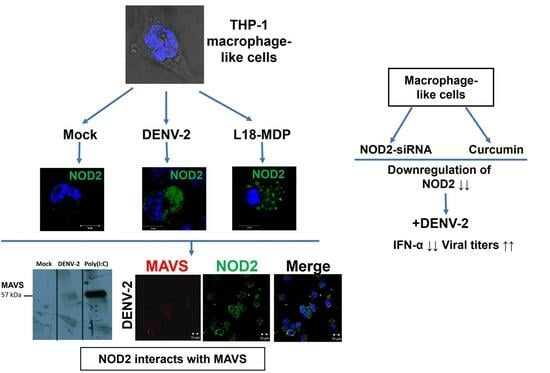
NOD2 Responds to Dengue Virus Type 2 Infection in Macrophage-like Cells Interacting with MAVS Adaptor and Affecting IFN-α Production and Virus Titers
Abstract
:1. Introduction
2. Materials and Methods
2.1. DENV-2 Strains, Culture, and Titration
2.2. THP-1 Cell Culture and Macrophage Differentiation
2.3. DENV-2 Infection in THP-1 Macrophage-like Cells
2.4. L18-MDP Treatment to Induce the Expression of NOD2
2.5. Poly I:C (PIC) Treatment to Induce MAVS Protein
2.6. Semi-Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR): mRNA Analysis
2.7. Western Blot (WB)
2.8. Confocal Microscopy
2.9. Co-Immunoprecipitation Assays (Co-IP)
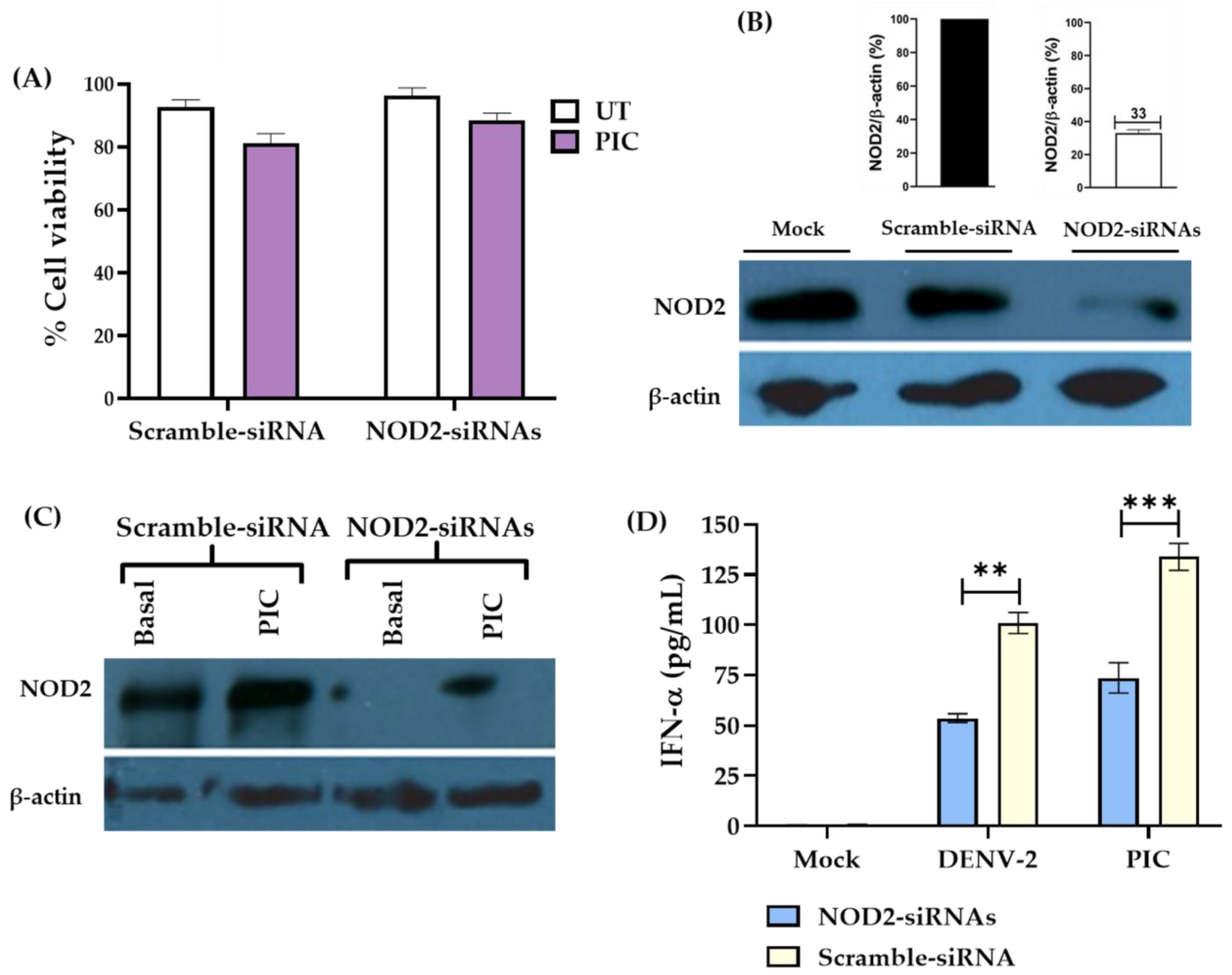
2.10. Small Interfering RNA (siRNA) Assays
2.11. IFN-α Quantification
2.12. Statistical Analysis
3. Results
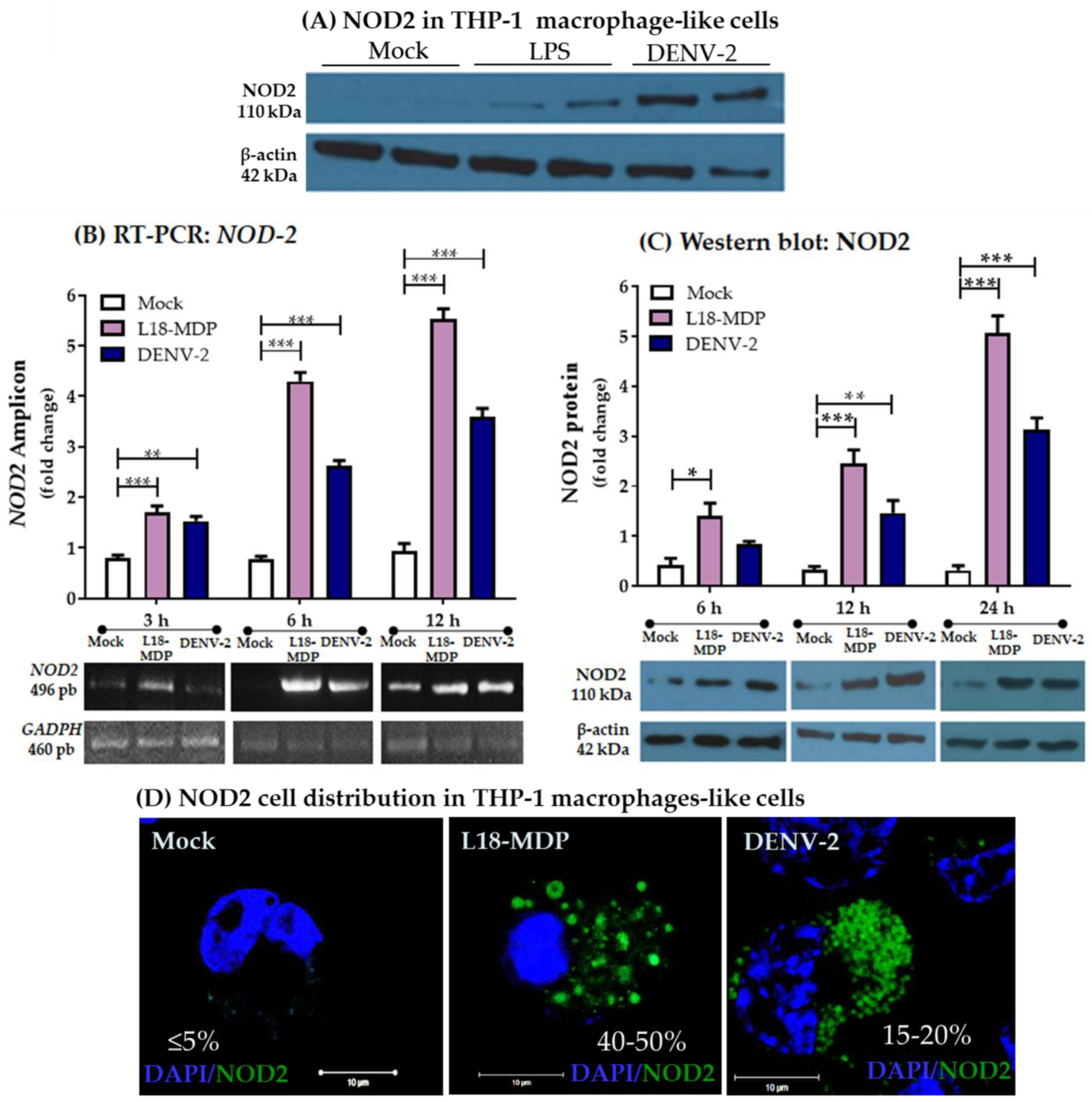
3.1. Infection with DENV-2 Upregulates NOD2 Expression in THP-1 Macrophage-like Cells
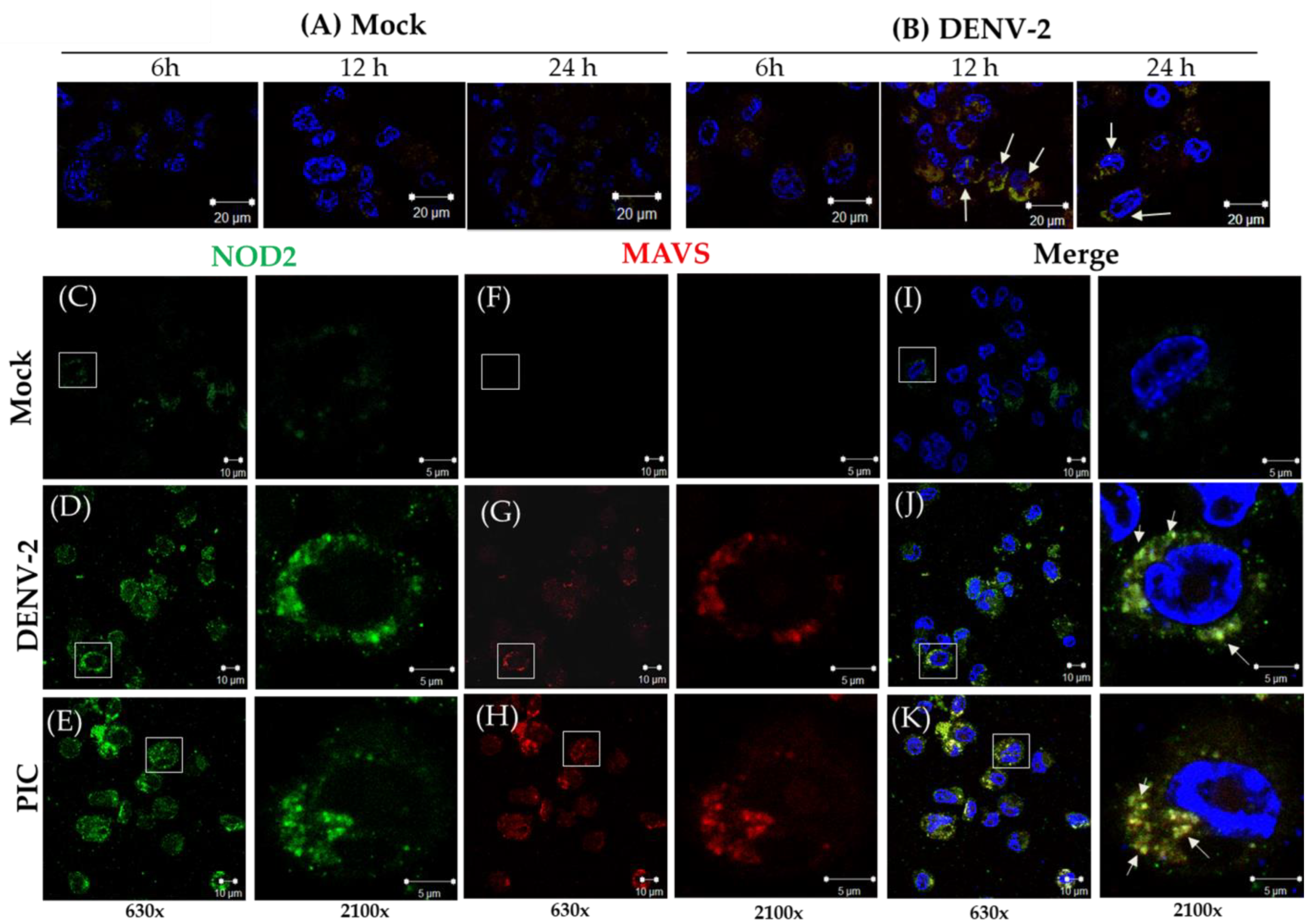
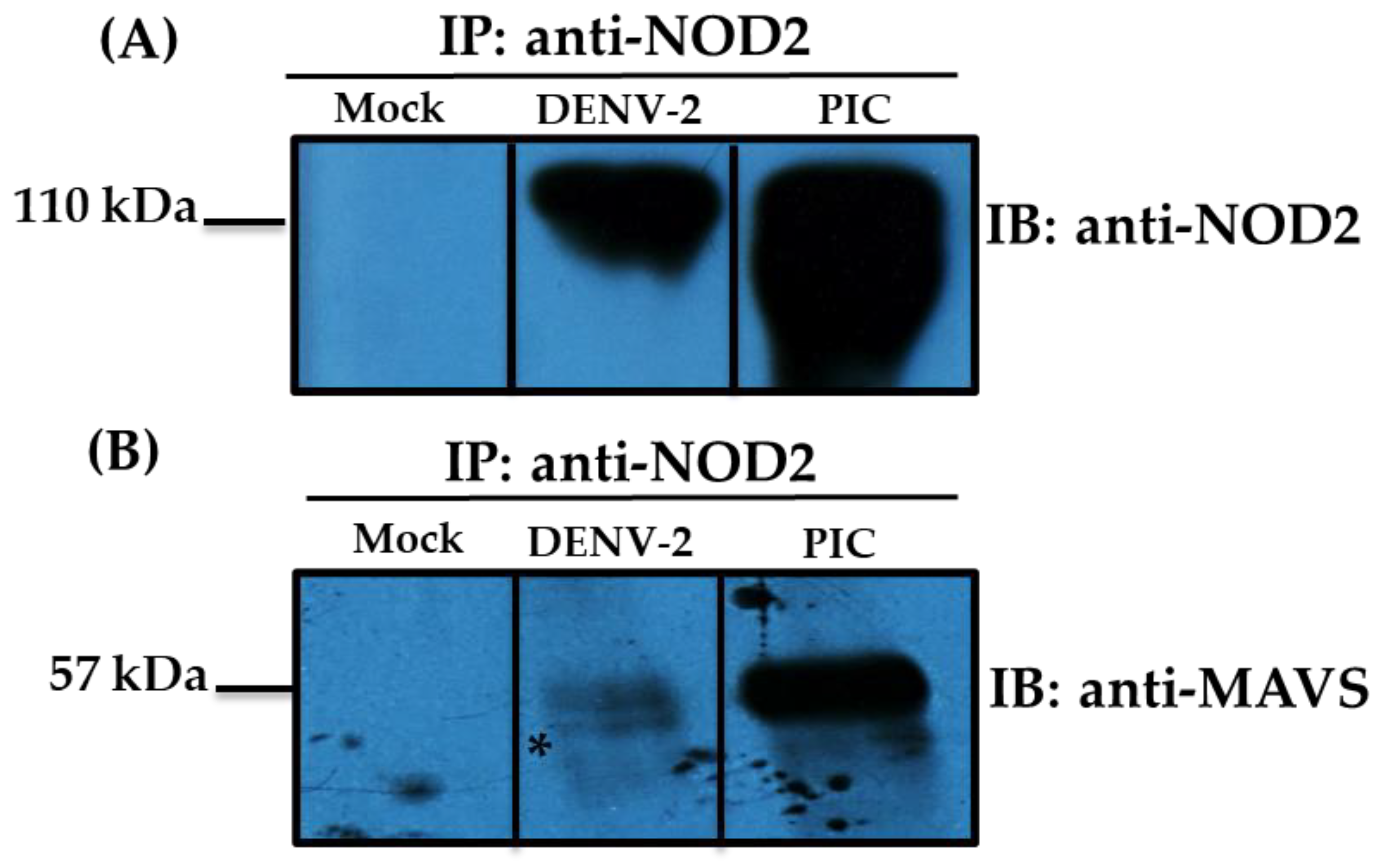
3.2. Detection of MAVS Effector and Interaction with NOD2 in THP-1 Macrophage-like Cells
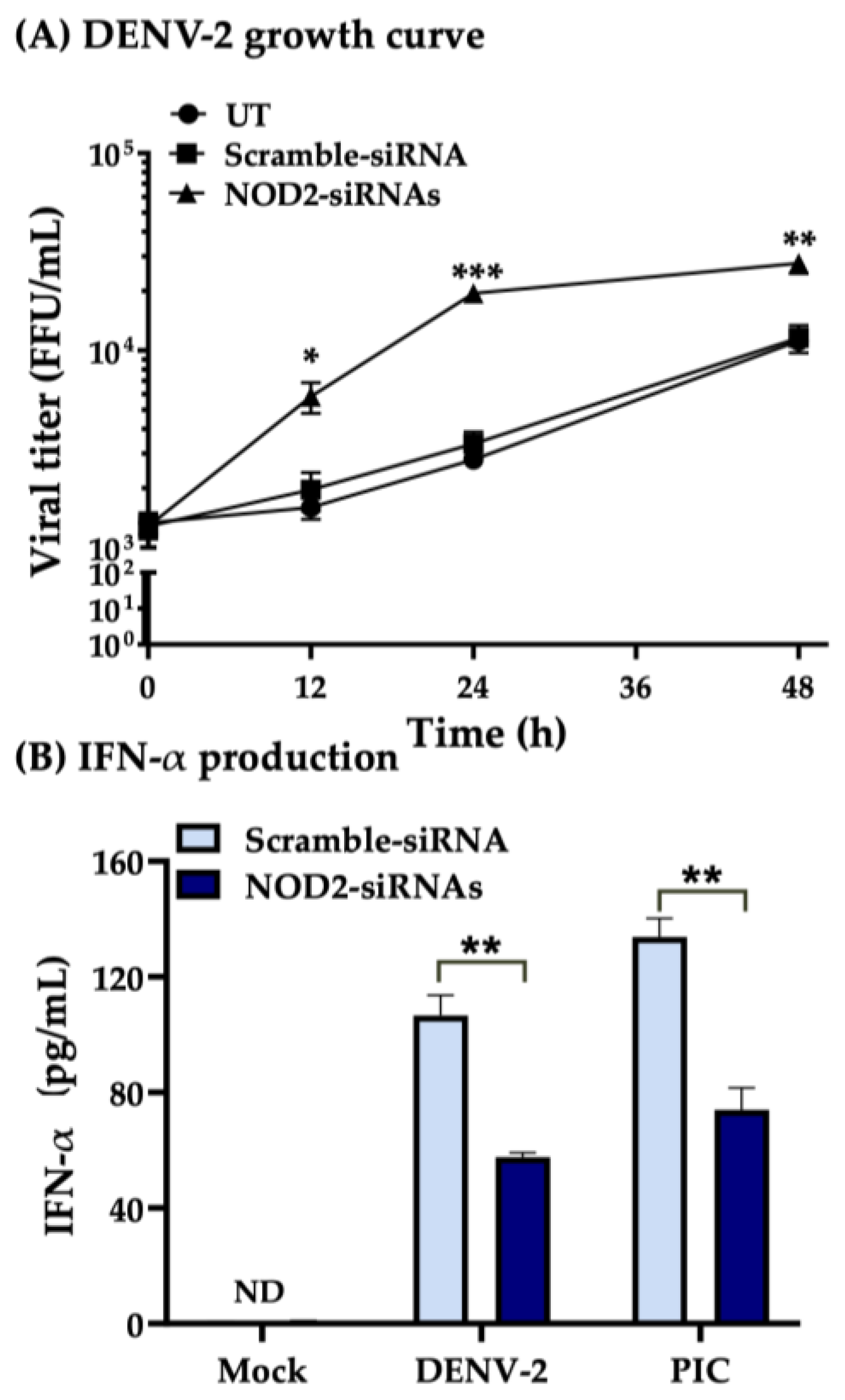
3.3. Downregulation of NOD2 Reduces the Production of IFN-α in Macrophage-like Cells Infected by DENV-2
3.4. NOD2 Inhibition Increased Viral Loads in THP-1 Macrophage-like Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The Global Distribution and Burden of Dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Rico-Hesse, R. Dengue Virus Virulence and Transmission Determinants. Curr. Top. Microbiol. Immunol. 2010, 338, 45–55. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Kuhn, R.J.; Rossmann, M.G. A Structural Perspective of the Flavivirus Life Cycle. Nat. Rev. Microbiol. 2005, 3, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.J.L.; Grouard-Vogel, G.; Sun, W.; Mascola, J.R.; Brachtel, E.; Putvatana, R.; Louder, M.K.; Filgueira, L.; Marovich, M.A.; Wong, H.K.; et al. Human Skin Langerhans Cells Are Targets of Dengue Virus Infection. Nat. Med. 2000, 6, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Marovich, M.; Grouard-Vogel, G.N.; Louder, M.; Eller, M.; Sun, W.; Wu, S.J.; Putvatana, R.; Murphy, G.; Tassaneetrithep, B.; Burgess, T.; et al. Human Dendritic Cells as Targets of Dengue Virus Infection. J. Investig. Dermatol. Symp. Proc. 2001, 6, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Jessie, K.; Fong, M.Y.; Devi, S.; Lam, S.K.; Wong, K.T. Localization of Dengue Virus in Naturally Infected Human Tissues, by Immunohistochemistry and in Situ Hybridization. J. Infect. Dis. 2004, 189, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Kwan, W.H.; Navarro-Sanchez, E.; Dumortier, H.; Decossas, M.; Vachon, H.; dos Santos, F.B.; Fridman, H.W.; Rey, F.A.; Harris, E.; Despres, P.; et al. Dermal-Type Macrophages Expressing CD209/DC-SIGN Show Inherent Resistance to Dengue Virus Growth. PLoS Neglected Trop. Dis. 2008, 2, e311. [Google Scholar] [CrossRef] [PubMed]
- Takaoka, A.; Yanai, H. Interferon Signalling Network in Innate Defence. Cell. Microbiol. 2006, 8, 907–922. [Google Scholar] [CrossRef] [PubMed]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I Interferons in Infectious Disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef]
- Wu, M.F.; Chen, S.T.; Yang, A.H.; Lin, W.W.; Lin, Y.L.; Chen, N.J.; Tsai, I.S.; Li, L.; Hsieh, S.L. CLEC5A Is Critical for Dengue Virus-Induced Inflammasome Activation in Human Macrophages. Blood 2013, 121, 95–106. [Google Scholar] [CrossRef]
- Hottz, E.D.; Lopes, J.F.; Freitas, C.; Valls-De-Souza, R.; Oliveira, M.F.; Bozza, M.T.; Da Poian, A.T.; Weyrich, A.S.; Zimmerman, G.A.; Bozza, F.A.; et al. Platelets Mediate Increased Endothelium Permeability in Dengue through NLRP3-Inflammasome Activation. Blood 2013, 122, 3405–3414. [Google Scholar] [CrossRef] [PubMed]
- Inohara, N.; Nuñez, G. NODs: Intracellular Proteins Involved in Inflammation and Apoptosis. Nat. Rev. Immunol. 2003, 3, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Moreira, L.O.; Zamboni, D.S. NOD1 and NOD2 Signaling in Infection and Inflammation. Front. Immunol. 2012, 3, 328. [Google Scholar] [CrossRef] [PubMed]
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of Innate Immune Antiviral Responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.G.; Park, J.H.; Reimer, T.; Baker, D.P.; Kawai, T.; Kumar, H.; Akira, S.; Wobus, C.; Núñez, G. Viral Infection Augments Nod1/2 Signaling to Potentiate Lethality Associated with Secondary Bacterial Infections. Cell Host Microbe 2011, 9, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Thomas, P.G.; Anand, P.K.; Vogel, P.; Milasta, S.; Martinez, J.; Huang, G.; Green, M.; Kundu, M.; Chi, H.; et al. Receptor Interacting Protein Kinase 2-Mediated Mitophagy Regulates Inflammasome Activation during Virus Infection. Nat. Immunol. 2013, 14, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Dugan, J.W.; Albor, A.; David, L.; Fowlkes, J.; Blackledge, M.T.; Martin, T.M.; Planck, S.R.; Rosenzweig, H.L.; Rosenbaum, J.T.; Davey, M.P. Nucleotide Oligomerization Domain-2 Interacts with 2′-5′-Oligoadenylate Synthetase Type 2 and Enhances RNase-L Function in THP-1 Cells. Mol. Immunol. 2009, 47, 560–566. [Google Scholar] [CrossRef]
- Olagnier, D.; Scholte, F.E.M.; Chiang, C.; Albulescu, I.C.; Nichols, C.; He, Z.; Lin, R.; Snijder, E.J.; van Hemert, M.J.; Hiscott, J. Inhibition of Dengue and Chikungunya Virus Infections by RIG-I-Mediated Type I Interferon-Independent Stimulation of the Innate Antiviral Response. J. Virol. 2014, 88, 4180–4194. [Google Scholar] [CrossRef]
- Lin, R.-J.; Yu, H.-P.; Chang, B.-L.; Tang, W.-C.; Liao, C.-L.; Lin, Y.-L. Distinct Antiviral Roles for Human 2′,5′-Oligoadenylate Synthetase Family Members against Dengue Virus Infection. J. Immunol. 2009, 183, 8035–8043. [Google Scholar] [CrossRef]
- Jadhav, N.J.; Gokhale, S.; Seervi, M.; Patil, P.S.; Alagarasu, K. Immunomodulatory Effect of 1, 25 Dihydroxy Vitamin D3 on the Expression of RNA Sensing Pattern Recognition Receptor Genes and Cytokine Response in Dengue Virus Infected U937-DC-SIGN Cells and THP-1 Macrophages. Int. Immunopharmacol. 2018, 62, 237–243. [Google Scholar] [CrossRef]
- Girardin, S.E.; Boneca, I.G.; Viala, J.; Chamaillard, M.; Labigne, A.; Thomas, G.; Philpott, D.J.; Sansonetti, P.J. Nod2 Is a General Sensor of Peptidoglycan through Muramyl Dipeptide (MDP) Detection. J. Biol. Chem. 2003, 278, 8869–8872. [Google Scholar] [CrossRef]
- Castañeda-Sánchez, J.I.; Domínguez-Martínez, D.A.; Olivar-Espinosa, N.; García-Pérez, B.E.; Loroño-Pino, M.A.; Luna-Herrera, J.; Salazar, M.I. Expression of Antimicrobial Peptides in Human Monocytic Cells and Neutrophils in Response to Dengue Virus Type 2. Intervirology 2016, 59, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Allen, I.C.; Moore, C.B.; Schneider, M.; Lei, Y.; Davis, B.K.; Scull, M.A.; Gris, D.; Roney, K.E.; Zimmermann, A.G.; Bowzard, J.B.; et al. NLRX1 Protein Attenuates Inflammatory Responses to Infection by Interfering with the RIG-I-MAVS and TRAF6-NF-ΚB Signaling Pathways. Immunity 2011, 34, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Liu, P.; Latz, E.; Golenbock, D.T.; Finberg, R.W.; Libraty, D.H. Flavivirus Activation of Plasmacytoid Dendritic Cells Delineates Key Elements of TLR7 Signaling beyond Endosomal Recognition. J. Immunol. 2006, 177, 7114–7121. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.T.; Chang, S.Y.; Lee, C.N.; Kao, C.L. Human TLR3 Recognizes Dengue Virus and Modulates Viral Replication in Vitro. Cell. Microbiol. 2009, 11, 604–615. [Google Scholar] [CrossRef]
- Nasirudeen, A.M.A.; Wong, H.H.; Thien, P.; Xu, S.; Lam, K.P.; Liu, D.X. RIG-I, MDA5 and TLR3 Synergistically Play an Important Role in Restriction of Dengue Virus Infection. PLoS Neglected Trop. Dis. 2011, 5, e926. [Google Scholar] [CrossRef]
- Nakamura, N.; Lill, J.R.; Phung, Q.; Jiang, Z.; Bakalarski, C.; De Mazière, A.; Klumperman, J.; Schlatter, M.; Delamarre, L.; Mellman, I. Endosomes Are Specialized Platforms for Bacterial Sensing and NOD2 Signalling. Nature 2014, 509, 240–244. [Google Scholar] [CrossRef]
- Ferrand, A.; Al Nabhani, Z.; Tapias, N.S.; Mas, E.; Hugot, J.P.; Barreau, F. NOD2 Expression in Intestinal Epithelial Cells Protects Toward the Development of Inflammation and Associated Carcinogenesis. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 357–369. [Google Scholar] [CrossRef]
- Negroni, A.; Pierdomenico, M.; Cucchiara, S.; Stronati, L. NOD2 and Inflammation: Current Insights. J. Inflamm. Res. 2018, 11, 49–60. [Google Scholar] [CrossRef]
- Domínguez-Martínez, D.A.; Núñez-Avellaneda, D.; Castañón-Sánchez, C.A.; Salazar, M.I. NOD2: Activation During Bacterial and Viral Infections, Polymorphisms and Potential as Therapeutic Target. Rev. Investig. Clínica 2018, 70, 18–28. [Google Scholar] [CrossRef]
- Bottermann, M.; James, L.C. Intracellular Antiviral Immunity. Adv. Virus Res. 2018, 100, 309–354. [Google Scholar] [CrossRef] [PubMed]
- Coutermarsh-Ott, S.; Eden, K.; Allen, I.C. Beyond the Inflammasome: Regulatory NOD-like Receptor Modulation of the Host Immune Response Following Virus Exposure. J. Gen. Virol. 2016, 97, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C. The Emerging Roles of NOD-like Receptors in Antiviral Innate Immune Signaling Pathways. Int. J. Biol. Macromol. 2021, 169, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Muharram, G.; Thépaut, M.; Lobert, P.E.; Grandjean, T.; Boulard, O.; Delacre, M.; Wakeford, E.; Wheeler, R.; Poulin, L.F.; Boneca, I.G.; et al. Activation of Nod2 Signaling upon Norovirus Infection Enhances Antiviral Immunity and Susceptibility to Colitis. Gut Microbes 2023, 15, 2249960. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domínguez-Martínez, D.A.; Pérez-Flores, M.S.; Núñez-Avellaneda, D.; Torres-Flores, J.M.; León-Avila, G.; García-Pérez, B.E.; Salazar, M.I. NOD2 Responds to Dengue Virus Type 2 Infection in Macrophage-like Cells Interacting with MAVS Adaptor and Affecting IFN-α Production and Virus Titers. Pathogens 2024, 13, 306. https://doi.org/10.3390/pathogens13040306
Domínguez-Martínez DA, Pérez-Flores MS, Núñez-Avellaneda D, Torres-Flores JM, León-Avila G, García-Pérez BE, Salazar MI. NOD2 Responds to Dengue Virus Type 2 Infection in Macrophage-like Cells Interacting with MAVS Adaptor and Affecting IFN-α Production and Virus Titers. Pathogens. 2024; 13(4):306. https://doi.org/10.3390/pathogens13040306
Chicago/Turabian StyleDomínguez-Martínez, Diana Alhelí, Mayra Silvia Pérez-Flores, Daniel Núñez-Avellaneda, Jesús M. Torres-Flores, Gloria León-Avila, Blanca Estela García-Pérez, and Ma Isabel Salazar. 2024. "NOD2 Responds to Dengue Virus Type 2 Infection in Macrophage-like Cells Interacting with MAVS Adaptor and Affecting IFN-α Production and Virus Titers" Pathogens 13, no. 4: 306. https://doi.org/10.3390/pathogens13040306