Cytomegalovirus Genetic Diversity and Evolution: Insights into Genotypes and Their Role in Viral Pathogenesis
Abstract
1. Introduction
2. Landscape of Genetic Diversity
2.1. HCMV Variability Is Mostly Found in “Islands” of Diversity
2.2. CMV Hypervariable Genes
2.2.1. Glycoprotein B
2.2.2. Glycoprotein N
2.2.3. Glycoprotein O
2.2.4. Glycoprotein H
2.3. Viral Cytokine/Chemokine Proteins (Human Cellular Homologues)
2.3.1. UL144
2.3.2. UL146 and UL147
2.4. Other Hypervariable Regions
2.5. Outside the Hypervariable Genes
2.6. Clinical Mutants with Nonfunctional Genes (Pseudogenes)
2.7. Repeats
3. Within Host Diversity
4. Clinical Significance of Multi-Allelic Regions
5. Evolution of Diversity
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Altevogt, P.; Sammar, M.; Hüser, L.; Kristiansen, G. Novel insights into the function of CD24: A driving force in cancer. Int. J. Cancer 2021, 148, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Aquino, V.H.; Figueiredo, L.T.M. High prevalence of renal transplant recipients infected with more than one cytomegalovirus glycoprotein B genotype. J. Med. Virol. 2000, 61, 138–142. [Google Scholar] [CrossRef]
- Arav-Boger, R. Strain Variation and Disease Severity in Congenital Cytomegalovirus Infection: In Search of a Viral Marker. Infect. Dis. Clin. 2015, 29, 401–414. [Google Scholar] [CrossRef]
- Arav-Boger, R.; Foster, C.B.; Zong, J.-C.; Pass, R.F. Human Cytomegalovirus–Encoded α-Chemokines Exhibit High Sequence Variability in Congenitally Infected Newborns. J. Infect. Dis. 2006, 193, 788–791. [Google Scholar] [CrossRef]
- Arav-Boger, R.; Willoughby, R.E.; Pass, R.F.; Zong, J.-C.; Jang, W.-J.; Alcendor, D.; Hayward, G.S. Polymorphisms of the Cytomegalovirus (CMV)–Encoded Tumor Necrosis Factor–α and β-Chemokine Receptors in Congenital CMV Disease. J. Infect. Dis. 2002, 186, 1057–1064. [Google Scholar] [CrossRef]
- Bale, J.F.; Petheram, S.J.; Robertson, M.; Murph, J.R.; Demmler, G. Human cytomegalovirus a sequence and UL144 variability in strains from infected children. J. Med. Virol. 2001, 65, 90–96. [Google Scholar] [CrossRef]
- Bale, J.F.; Petheram, S.J.; Souza, I.E.; Murph, J.R. Cytomegalovirus reinfection in young children. J. Pediatr. 1996, 128, 347–352. [Google Scholar] [CrossRef]
- Barbi, M.; Binda, S.; Caroppo, S.; Primache, V.; Didò, P.; Guidotti, P.; Corbetta, C.; Melotti, D. CMV gB genotypes and outcome of vertical transmission: Study on dried blood spots of congenitally infected babies. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2001, 21, 75–79. [Google Scholar] [CrossRef]
- Bates, M.; Monze, M.; Bima, H.; Kapambwe, M.; Kasolo, F.C.; Gompels, U.A. High human cytomegalovirus loads and diverse linked variable genotypes in both HIV-1 infected and exposed, but uninfected, children in Africa. Virology 2008, 382, 28–36. [Google Scholar] [CrossRef]
- Benedict, C.A.; Butrovich, K.D.; Lurain, N.S.; Corbeil, J.; Rooney, I.; Schneider, P.; Tschopp, J.; Ware, C.F. Cutting edge: A novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J. Immunol. 1999, 162, 6967–6970. [Google Scholar] [CrossRef]
- Beninga, J.; Kalbacher, H.; Mach, M. Analysis of T Helper Cell Response to Glycoprotein H (gpUL75) of Human Cytomegalovirus: Evidence for Strain-Specific T Cell Determinants. J. Infect. Dis. 1996, 173, 1051–1061. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Berg, C.; Rosenkilde, M.M.; Benfield, T.; Nielsen, L.; Sundelin, T.; Lüttichau, H.R. The frequency of cytomegalovirus non-ELR UL146 genotypes in neonates with congenital CMV disease is comparable to strains in the background population. BMC Infect. Dis. 2021, 21, 386. [Google Scholar] [CrossRef] [PubMed]
- Boeckh, M.; Geballe, A.P. Cytomegalovirus: Pathogen, paradigm, and puzzle. J. Clin. Investig. 2011, 121, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, C.M.; Cardin, R.D. Don’t Go Breaking My Heart: MCMV as a Model for HCMV-Associated Cardiovascular Diseases. Pathogens 2021, 10, 619. [Google Scholar] [CrossRef]
- Boppana, S.B.; Ross, S.A.; Fowler, K.B. Congenital Cytomegalovirus Infection: Clinical Outcome. Clin. Infect. Dis. 2013, 57 (Suppl. S4), S178–S181. [Google Scholar] [CrossRef]
- Bradley, A.J.; Kovács, I.J.; Gatherer, D.; Dargan, D.J.; Alkharsah, K.R.; Chan, P.K.S.; Carman, W.F.; Dedicoat, M.; Emery, V.C.; Geddes, C.C.; et al. Genotypic analysis of two hypervariable human cytomegalovirus genes. J. Med. Virol. 2008, 80, 1615–1623. [Google Scholar] [CrossRef]
- Brait, N.; Stögerer, T.; Kalser, J.; Adler, B.; Kunz, I.; Benesch, M.; Kropff, B.; Mach, M.; Puchhammer-Stöckl, E.; Görzer, I. Influence of Human Cytomegalovirus Glycoprotein O Polymorphism on the Inhibitory Effect of Soluble Forms of Trimer- and Pentamer-Specific Entry Receptors. J. Virol. 2020, 94, e00107-20. [Google Scholar] [CrossRef]
- Britt, W.J. Congenital Human Cytomegalovirus Infection and the Enigma of Maternal Immunity. J. Virol. 2017, 91, e02392-16. [Google Scholar] [CrossRef]
- Burkhardt, C.; Himmelein, S.; Britt, W.; Winkler, T.; Mach, M. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J. Gen. Virol. 2009, 90, 1951–1961. [Google Scholar] [CrossRef]
- Cha, T.A.; Tom, E.; Kemble, G.W.; Duke, G.M.; Mocarski, E.S.; Spaete, R.R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 1996, 70, 78–83. [Google Scholar] [CrossRef]
- Chandler, S.H.; Handsfield, H.H.; McDougall, J.K. Isolation of Multiple Strains of Cytomegalovirus from Women Attending a Clinic for Sexually Transmitted Diseases. J. Infect. Dis. 1987, 155, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Travers, P.; Walport, M.; Shlomchik, M.J. The major histocompatibility complex and its functions. In Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science: New York City, NY, USA, 2001. Available online: https://www.ncbi.nlm.nih.gov/books/NBK27156/ (accessed on 26 July 2024).
- Charles, O.J.; Venturini, C.; Breuer, J. cmvdrg—An R package for Human Cytomegalovirus Antiviral Drug Resistance Genotyping. bioRxiv 2020. [Google Scholar] [CrossRef]
- Charles, O.J.; Venturini, C.; Gantt, S.; Atkinson, C.; Griffiths, P.; Goldstein, R.A.; Breuer, J. Genomic and geographical structure of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 2023, 120, e2221797120. [Google Scholar] [CrossRef] [PubMed]
- Chee, M.S.; Bankier, A.T.; Beck, S.; Bohni, R.; Brown, C.M.; Cerny, R.; Horsnell, T.; Hutchison, C.A.; Kouzarides, T.; Martignetti, J.A.; et al. Analysis of the Protein-Coding Content of the Sequence of Human Cytomegalovirus Strain AD169. In Cytomegaloviruses; McDougall, J.K., Ed.; Springer: Berlin/Heidelberg, Germany, 1990; pp. 125–169. [Google Scholar] [CrossRef]
- Chou, S. Reactivation and Recombination of Multiple Cytomegalovirus Strains from Individual Organ Donors. J. Infect. Dis. 1989, 160, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Chou, S. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 1992, 188, 388–390. [Google Scholar] [CrossRef]
- Chou, S. Diverse Cytomegalovirus UL27 Mutations Adapt to Loss of Viral UL97 Kinase Activity under Maribavir. Antimicrob. Agents Chemother. 2009, 53, 81–85. [Google Scholar] [CrossRef]
- Chou, S.; Dennison, K.M. Analysis of Interstrain Variation in Cytomegalovirus Glycoprotein B Sequences Encoding Neutralization-Related Epitopes. J. Infect. Dis. 1991, 163, 1229–1234. [Google Scholar] [CrossRef]
- Coaquette, A.; Bourgeois, A.; Dirand, C.; Varin, A.; Chen, W.; Herbein, G. Mixed Cytomegalovirus Glycoprotein B Genotypes in Immunocompromised Patients. Clin. Infect. Dis. 2004, 39, 155–161. [Google Scholar] [CrossRef]
- Correia, S.; Bridges, R.; Wegner, F.; Venturini, C.; Palser, A.; Middeldorp, J.M.; Cohen, J.I.; Lorenzetti, M.A.; Bassano, I.; White, R.E.; et al. Sequence Variation of Epstein-Barr Virus: Viral Types, Geography, Codon Usage, and Diseases. J. Virol. 2018, 92, e01132-18. [Google Scholar] [CrossRef]
- Cruz, D.V.; Nelson, C.S.; Tran, D.; Barry, P.A.; Kaur, A.; Koelle, K.; Permar, S.R. Intrahost cytomegalovirus population genetics following antibody pretreatment in a monkey model of congenital transmission. PLoS Pathog. 2020, 16, e1007968. [Google Scholar] [CrossRef]
- Cudini, J.; Roy, S.; Houldcroft, C.J.; Bryant, J.M.; Depledge, D.P.; Tutill, H.; Veys, P.; Williams, R.; Worth, A.J.J.; Tamuri, A.U.; et al. Human cytomegalovirus haplotype reconstruction reveals high diversity due to superinfection and evidence of within-host recombination. Proc. Natl. Acad. Sci. USA 2019, 116, 5693–5698. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Freed, D.C.; Wang, D.; Qiu, P.; Li, F.; Fu, T.-M.; Kauvar, L.M.; McVoy, M.A. Impact of Antibodies and Strain Polymorphisms on Cytomegalovirus Entry and Spread in Fibroblasts and Epithelial Cells. J. Virol. 2017, 91, e01650-16. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.; Gatherer, D.; Hilfrich, B.; Baluchova, K.; Dargan, D.J.; Thomson, M.; Griffiths, P.D.; Wilkinson, G.W.G.; Schulz, T.F.; Davison, A.J. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 2010, 91, 605–615. [Google Scholar] [CrossRef]
- Dargan, D.J.; Douglas, E.; Cunningham, C.; Jamieson, F.; Stanton, R.J.; Baluchova, K.; McSharry, B.P.; Tomasec, P.; Emery, V.C.; Percivalle, E.; et al. Sequential mutations associated with adaptation of human cytomegalovirus to growth in cell culture. J. Gen. Virol. 2010, 91, 1535–1546. [Google Scholar] [CrossRef]
- Davis, C.L.; Field, D.; Metzgar, D.; Saiz, R.; Morin, P.A.; Smith, I.L.; Spector, S.A.; Wills, C. Numerous Length Polymorphisms at Short Tandem Repeats in Human Cytomegalovirus. J. Virol. 1999, 73, 6265–6270. [Google Scholar] [CrossRef]
- Davison, A.J.; Dolan, A.; Akter, P.; Addison, C.; Dargan, D.J.; Alcendor, D.J.; McGeoch, D.J.; Hayward, G.S. The human cytomegalovirus genome revisited: Comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 2003, 84 Pt 1, 17–28. [Google Scholar] [CrossRef]
- Day, L.Z.; Stegmann, C.; Schultz, E.P.; Lanchy, J.-M.; Yu, Q.; Ryckman, B.J. Polymorphisms in Human Cytomegalovirus Glycoprotein O (gO) Exert Epistatic Influences on Cell-Free and Cell-to-Cell Spread and Antibody Neutralization on gH Epitopes. J. Virol. 2020, 94, e02051-19. [Google Scholar] [CrossRef]
- Depledge, D.P.; Palser, A.L.; Watson, S.J.; Lai, I.Y.-C.; Gray, E.R.; Grant, P.; Kanda, R.K.; Leproust, E.; Kellam, P.; Breuer, J. Specific Capture and Whole-Genome Sequencing of Viruses from Clinical Samples. PLoS ONE 2011, 6, e27805. [Google Scholar] [CrossRef]
- Dhingra, A.; Götting, J.; Varanasi, P.R.; Steinbrueck, L.; Camiolo, S.; Zischke, J.; Heim, A.; Schulz, T.F.; Weissinger, E.M.; Kay-Fedorov, P.C.; et al. Human cytomegalovirus multiple-strain infections and viral population diversity in haematopoietic stem cell transplant recipients analysed by high-throughput sequencing. Med. Microbiol. Immunol. 2021, 210, 291–304. [Google Scholar] [CrossRef]
- Dobbins, G.C.; Patki, A.; Chen, D.; Tiwari, H.K.; Hendrickson, C.; Britt, W.J.; Fowler, K.; Chen, J.Y.; Boppana, S.B.; Ross, S.A. Association of CMV genomic mutations with symptomatic infection and hearing loss in congenital CMV infection. BMC Infect. Dis. 2019, 19, 1046. [Google Scholar] [CrossRef]
- Dolan, A.; Cunningham, C.; Hector, R.D.; Hassan-Walker, A.F.; Lee, L.; Addison, C.; Dargan, D.J.; McGeoch, D.J.; Gatherer, D.; Emery, V.C.; et al. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 2004, 85, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Cao, L.; Zheng, D.; Su, L.; Lu, L.; Dong, Z.; Xu, M.; Xu, J. Distribution of CMV envelope glycoprotein B, H and N genotypes in infants with congenital cytomegalovirus symptomatic infection. Front. Pediatr. 2023, 11, 1112645. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Zheng, P.; Tang, J.; Liu, Y. CD24: From A to Z. Cell. Mol. Immunol. 2010, 7, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Faure-Della Corte, M.; Samot, J.; Garrigue, I.; Magnin, N.; Reigadas, S.; Couzi, L.; Dromer, C.; Velly, J.-F.; Déchanet-Merville, J.; Fleury, H.J.A.; et al. Variability and recombination of clinical human cytomegalovirus strains from transplantation recipients. J. Clin. Virol. 2010, 47, 161–169. [Google Scholar] [CrossRef]
- Freed, D.C.; Tang, Q.; Tang, A.; Li, F.; He, X.; Huang, Z.; Meng, W.; Xia, L.; Finnefrock, A.C.; Durr, E.; et al. Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proc. Natl. Acad. Sci. USA 2013, 110, E4997–E5005. [Google Scholar] [CrossRef]
- Fulkerson, H.L.; Nogalski, M.T.; Collins-McMillen, D.; Yurochko, A.D. Overview of Human Cytomegalovirus Pathogenesis. In Human Cytomegaloviruses: Methods and Protocols; Yurochko, A.D., Ed.; Springer: New York, NY, USA, 2021; pp. 1–18. [Google Scholar] [CrossRef]
- Garrigue, I.; Corte, M.F.-D.; Magnin, N.; Recordon-Pinson, P.; Couzi, L.; Lebrette, M.-E.; Schrive, M.-H.; Roncin, L.; Taupin, J.-L.; Déchanet-Merville, J.; et al. UL40 Human Cytomegalovirus Variability Evolution Patterns Over Time in Renal Transplant Recipients. Transplantation 2008, 86, 826. [Google Scholar] [CrossRef]
- Görzer, I.; Guelly, C.; Trajanoski, S.; Puchhammer-Stöckl, E. Deep Sequencing Reveals Highly Complex Dynamics of Human Cytomegalovirus Genotypes in Transplant Patients over Time. J. Virol. 2010, 84, 7195–7203. [Google Scholar] [CrossRef]
- Görzer, I.; Kerschner, H.; Jaksch, P.; Bauer, C.; Seebacher, G.; Klepetko, W.; Puchhammer-Stöckl, E. Virus load dynamics of individual CMV-genotypes in lung transplant recipients with mixed-genotype infections. J. Med. Virol. 2008, 80, 1405–1414. [Google Scholar] [CrossRef]
- Gretch, D.R.; Kari, B.; Rasmussen, L.; Gehrz, R.C.; Stinski, M.F. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J. Virol. 1988, 62, 875–881. [Google Scholar] [CrossRef]
- Grosjean, J.; Hantz, S.; Cotin, S.; Baclet, M.C.; Mengelle, C.; Trapes, L.; Virey, B.; Undreiner, F.; Brosset, P.; Pasquier, C.; et al. Direct genotyping of cytomegalovirus envelope glycoproteins from toddler’s saliva samples. J. Clin. Virol. 2009, 46, S43–S48. [Google Scholar] [CrossRef]
- Gugliesi, F.; Pasquero, S.; Griffante, G.; Scutera, S.; Albano, C.; Pacheco, S.F.C.; Riva, G.; Dell’Oste, V.; Biolatti, M. Human Cytomegalovirus and Autoimmune Diseases: Where Are We? Viruses 2021, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Zhang, L.; Ye, S.; Hu, Y.; Li, B.; Sun, X.; Mao, C.; Xu, J.; Chen, Y.; Zhang, L.; et al. Polymorphisms and features of cytomegalovirus UL144 and UL146 in congenitally infected neonates with hepatic involvement. PLoS ONE 2017, 12, e0171959. [Google Scholar] [CrossRef] [PubMed]
- Haberland, M.; Meyer-König, U.; Hufert, F.T. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J. Gen. Virol. 1999, 80, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Hage, E.; Wilkie, G.S.; Linnenweber-Held, S.; Dhingra, A.; Suárez, N.M.; Schmidt, J.J.; Kay-Fedorov, P.C.; Mischak-Weissinger, E.; Heim, A.; Schwarz, A.; et al. Characterization of Human Cytomegalovirus Genome Diversity in Immunocompromised Hosts by Whole-Genome Sequencing Directly From Clinical Specimens. J. Infect. Dis. 2017, 215, 1673–1683. [Google Scholar] [CrossRef]
- Hansen, S.G.; Powers, C.J.; Richards, R.; Ventura, A.B.; Ford, J.C.; Siess, D.; Axthelm, M.K.; Nelson, J.A.; Jarvis, M.A.; Picker, L.J.; et al. Evasion of CD8+ T Cells Is Critical for Superinfection by Cytomegalovirus. Science 2010, 328, 102–106. [Google Scholar] [CrossRef]
- Hassan-Walker, A.F.; Okwuadi, S.; Lee, L.; Griffiths, P.D.; Emery, V.C. Sequence variability of the α-chemokine UL146 from clinical strains of human cytomegalovirus. J. Med. Virol. 2004, 74, 573–579. [Google Scholar] [CrossRef]
- Heo, J.; Petheram, S.; Demmler, G.; Murph, J.R.; Adler, S.P.; Bale, J.; Sparer, T.E. Polymorphisms within human cytomegalovirus chemokine (UL146/UL147) and cytokine receptor genes (UL144) are not predictive of sequelae in congenitally infected children. Virology 2008, 378, 86. [Google Scholar] [CrossRef]
- Houldcroft, C.J.; Beale, M.A.; Breuer, J. Clinical and biological insights from viral genome sequencing. Nat. Rev. Microbiol. 2017, 15, 183–192. [Google Scholar] [CrossRef]
- Isaacson, M.K.; Compton, T. Human Cytomegalovirus Glycoprotein B Is Required for Virus Entry and Cell-to-Cell Spread but Not for Virion Attachment, Assembly, or Egress. J. Virol. 2009, 83, 3891–3903. [Google Scholar] [CrossRef]
- Ishibashi, K.; Tokumoto, T.; Shirakawa, H.; Hashimoto, K.; Kushida, N.; Yanagida, T.; Shishido, K.; Aikawa, K.; Yamaguchi, O.; Toma, H.; et al. Association between antibody response against cytomegalovirus strain-specific glycoprotein H epitopes and HLA-DR. Microbiol. Immunol. 2009, 53, 412–416. [Google Scholar] [CrossRef]
- Kalser, J.; Adler, B.; Mach, M.; Kropff, B.; Puchhammer-Stöckl, E.; Görzer, I. Differences in Growth Properties among Two Human Cytomegalovirus Glycoprotein O Genotypes. Front. Microbiol. 2017, 8, 1609. [Google Scholar] [CrossRef] [PubMed]
- Kaufer, B.B.; Jarosinski, K.W.; Osterrieder, N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J. Exp. Med. 2011, 208, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Lassalle, F.; Depledge, D.P.; Reeves, M.B.; Brown, A.C.; Christiansen, M.T.; Tutill, H.J.; Williams, R.J.; Einer-Jensen, K.; Holdstock, J.; Atkinson, C.; et al. Islands of linkage in an ocean of pervasive recombination reveals two-speed evolution of human cytomegalovirus genomes. Virus Evol. 2016, 2, vew017. [Google Scholar] [CrossRef]
- Lurain, N.S.; Chou, S. Antiviral Drug Resistance of Human Cytomegalovirus. Clin. Microbiol. Rev. 2010, 23, 689–712. [Google Scholar] [CrossRef]
- Lurain, N.S.; Fox, A.M.; Lichy, H.M.; Bhorade, S.M.; Ware, C.F.; Huang, D.D.; Kwan, S.-P.; Garrity, E.R.; Chou, S. Analysis of the human cytomegalovirus genomic region from UL146 through UL147A reveals sequence hypervariability, genotypic stability, and overlapping transcripts. Virol. J. 2006, 3, 4. [Google Scholar] [CrossRef]
- Lurain, N.S.; Kapell, K.S.; Huang, D.D.; Short, J.A.; Paintsil, J.; Winkfield, E.; Benedict, C.A.; Ware, C.F.; Bremer, J.W. Human cytomegalovirus UL144 open reading frame: Sequence hypervariability in low-passage clinical isolates. J. Virol. 1999, 73, 10040–10050. [Google Scholar] [CrossRef]
- Mach, M.; Kropff, B.; Dal Monte, P.; Britt, W. Complex Formation by Human Cytomegalovirus Glycoproteins M (gpUL100) and N (gpUL73). J. Virol. 2000, 74, 11881–11892. [Google Scholar] [CrossRef]
- Manicklal, S.; Emery, V.C.; Lazzarotto, T.; Boppana, S.B.; Gupta, R.K. The ‘silent’ global burden of congenital cytomegalovirus. Clin. Microbiol. Rev. 2013, 26, 86–102. [Google Scholar] [CrossRef]
- Manuel, O.; Åsberg, A.; Pang, X.; Rollag, H.; Emery, V.C.; Preiksaitis, J.K.; Kumar, D.; Pescovitz, M.D.; Bignamini, A.A.; Hartmann, A.; et al. Impact of Genetic Polymorphisms in Cytomegalovirus Glycoprotein B on Outcomes in Solid-Organ Transplant Recipients with Cytomegalovirus Disease. Clin. Infect. Dis. 2009, 49, 1160–1166. [Google Scholar] [CrossRef]
- Manuel, O.; Pang, X.L.; Humar, A.; Kumar, D.; Doucette, K.; Preiksaitis, J.K. An Assessment of Donor-to-Recipient Transmission Patterns of Human Cytomegalovirus by Analysis of Viral Genomic Variants. J. Infect. Dis. 2009, 199, 1621–1628. [Google Scholar] [CrossRef]
- Masse, M.J.; Karlin, S.; Schachtel, G.A.; Mocarski, E.S. Human cytomegalovirus origin of DNA replication (oriLyt) resides within a highly complex repetitive region. Proc. Natl. Acad. Sci. USA 1992, 89, 5246–5250. [Google Scholar] [CrossRef] [PubMed]
- Mattick, C.; Dewin, D.; Polley, S.; Sevilla-Reyes, E.; Pignatelli, S.; Rawlinson, W.; Wilkinson, G.; Dal Monte, P.; Gompels, U.A. Linkage of human cytomegalovirus glycoprotein gO variant groups identified from worldwide clinical isolates with gN genotypes, implications for disease associations and evidence for N-terminal sites of positive selection. Virology 2004, 318, 582–597. [Google Scholar] [CrossRef] [PubMed]
- McFaline-Figueroa, J.R.; Wen, P.Y. The Viral Connection to Glioblastoma. Curr. Infect. Dis. Rep. 2017, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- McGeoch, D.J.; Rixon, F.J.; Davison, A.J. Topics in herpesvirus genomics and evolution. Virus Res. 2006, 117, 90–104. [Google Scholar] [CrossRef] [PubMed]
- McSharry, B.P.; Avdic, S.; Slobedman, B. Human cytomegalovirus encoded homologs of cytokines, chemokines and their receptors: Roles in immunomodulation. Viruses 2012, 4, 2448–2470. [Google Scholar] [CrossRef]
- Meyer-König, U.; Ebert, K.; Schrage, B.; Pollak, S.; Hufert, F.T. Simultaneous infection of healthy people with multiple human cytomegalovirus strains. Lancet 1998, 352, 1280–1281. [Google Scholar] [CrossRef]
- Meyer-König, U.; Vogelberg, C.; Bongarts, A.; Kampa, D.; Delbrück, R.; Wolff-Vorbeck, G.; Kirste, G.; Haberland, M.; Hufert, F.T.; von Laer, D. Glycoprotein B genotype correlates with cell tropism in vivo of human cytomegalovirus infection. J. Med. Virol. 1998, 55, 75–81. [Google Scholar] [CrossRef]
- Michaelis, M.; Doerr, H.W.; Cinatl, J. The Story of Human Cytomegalovirus and Cancer: Increasing Evidence and Open Questions. Neoplasia 2009, 11, 1–9. [Google Scholar] [CrossRef]
- Murphy, E.; Shenk, T.E. Human Cytomegalovirus Genome. In Human Cytomegalovirus; Shenk, T.E., Stinski, M.F., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–19. [Google Scholar] [CrossRef]
- Nanamiya, H.; Tanaka, D.; Hiyama, G.; Isogai, T.; Watanabe, S. Detection of four isomers of the human cytomegalovirus genome using nanopore long-read sequencing. Virus Genes 2024, 60, 377–384. [Google Scholar] [CrossRef]
- Nelson, C.S.; Huffman, T.; Jenks, J.A.; Cisneros de la Rosa, E.; Xie, G.; Vandergrift, N.; Pass, R.F.; Pollara, J.; Permar, S.R. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc. Natl. Acad. Sci. USA 2018, 115, 6267–6272. [Google Scholar] [CrossRef]
- Nijman, J.; Mandemaker, F.S.; Verboon-Maciolek, M.A.; Aitken, S.C.; van Loon, A.M.; de Vries, L.S.; Schuurman, R. Genotype Distribution, Viral Load and Clinical Characteristics of Infants with Postnatal or Congenital Cytomegalovirus Infection. PLoS ONE 2014, 9, e108018. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Novak, Z.; Ross, S.A.; Patro, R.K.; Pati, S.K.; Kumbla, R.A.; Brice, S.; Boppana, S.B. Cytomegalovirus Strain Diversity in Seropositive Women. J. Clin. Microbiol. 2008, 46, 882–886. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Slyker, J.A.; Roy, S.; Bryant, J.; Atkinson, C.; Cudini, J.; Farquhar, C.; Griffiths, P.; Kiarie, J.; Morfopoulou, S.; et al. Mixed cytomegalovirus genotypes in HIV-positive mothers show compartmentalization and distinct patterns of transmission to infants. eLife 2020, 9, e63199. [Google Scholar] [CrossRef] [PubMed]
- Paradowska, E.; Jabłońska, A.; Płóciennikowska, A.; Studzińska, M.; Suski, P.; Wiśniewska-Ligier, M.; Dzierżanowska-Fangrat, K.; Kasztelewicz, B.; Woźniakowska-Gęsicka, T.; Leśnikowski, Z.J. Cytomegalovirus alpha-chemokine genotypes are associated with clinical manifestations in children with congenital or postnatal infections. Virology 2014, 462–463, 207–217. [Google Scholar] [CrossRef]
- Pati, S.; Pinninti, S.; Novak, Z.; Chowdhury, N.; Patro, R.; Fowler, K.; Ross, S.; Boppana, S. Genotypic Diversity and Mixed Infection in Newborn Disease and Hearing Loss in Congenital Cytomegalovirus Infection. Pediatr. Infect. Dis. J. 2013, 32, 1050–1054. [Google Scholar] [CrossRef]
- Penfold, M.E.T.; Dairaghi, D.J.; Duke, G.M.; Saederup, N.; Mocarski, E.S.; Kemble, G.W.; Schall, T.J. Cytomegalovirus encodes a potent α chemokine. Proc. Natl. Acad. Sci. USA 1999, 96, 9839–9844. [Google Scholar] [CrossRef]
- Perdue, M.L.; García, M.; Senne, D.; Fraire, M. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997, 49, 173–186. [Google Scholar] [CrossRef]
- Picone, O.; Costa, J.-M.; Ville, Y.; Chaix, M.-L.; Rouzioux, C.; Leruez-Ville, M. Genetic polymorphism of cytomegalovirus strains responsible of congenital infections. Pathol.-Biol. 2004, 52, 534–539. [Google Scholar] [CrossRef]
- Pignatelli, S.; Dal Monte, P.; Landini, M.P. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J. Gen. Virol. 2001, 82, 2777–2784. [Google Scholar] [CrossRef]
- Pignatelli, S.; Dal Monte, P.; Rossini, G.; Chou, S.; Gojobori, T.; Hanada, K.; Guo, J.J.; Rawlinson, W.; Britt, W.; Mach, M.; et al. Human cytomegalovirus glycoprotein N (gpUL73-gN) genomic variants: Identification of a novel subgroup, geographical distribution and evidence of positive selective pressure. J. Gen. Virol. 2003, 84, 647–655. [Google Scholar] [CrossRef]
- Pignatelli, S.; Lazzarotto, T.; Gatto, M.R.; Dal Monte, P.; Landini, M.P.; Faldella, G.; Lanari, M. Cytomegalovirus gN Genotypes Distribution among Congenitally Infected Newborns and Their Relationship with Symptoms at Birth and Sequelae. Clin. Infect. Dis. 2010, 51, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, S.; Monte, P.D.; Rossini, G.; Landini, M.P. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev. Med. Virol. 2004, 14, 383–410. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, S.A.; Starr, S.E.; Friedman, H.M.; Gönczöl, E.; Weibel, R.E. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 1989, 159, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Prichard, M.N.; Penfold, M.E.T.; Duke, G.M.; Spaete, R.R.; Kemble, G.W. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 2001, 11, 191–200. [Google Scholar] [CrossRef]
- Prod’homme, V.; Tomasec, P.; Cunningham, C.; Lemberg, M.K.; Stanton, R.J.; McSharry, B.P.; Wang, E.C.Y.; Cuff, S.; Martoglio, B.; Davison, A.J.; et al. Human cytomegalovirus UL40 signal peptide regulates cell surface expression of the NK cell ligands HLA-E and gpUL18. J. Immunol. 2012, 188, 2794–2804. [Google Scholar] [CrossRef]
- Puchhammer-Stöckl, E.; Görzer, I. Cytomegalovirus and Epstein-Barr virus subtypes—The search for clinical significance. J. Clin. Virol. 2006, 36, 239–248. [Google Scholar] [CrossRef]
- Puchhammer-Stöckl, E.; Görzer, I. Human cytomegalovirus: An enormous variety of strains and their possible clinical significance in the human host. Future Virol. 2011, 6, 259–271. [Google Scholar] [CrossRef]
- Qi, Y.; Mao, Z.-Q.; Ruan, Q.; He, R.; Ma, Y.-P.; Sun, Z.-R.; Ji, Y.-H.; Huang, Y. Human cytomegalovirus (HCMV) UL139 open reading frame: Sequence variants are clustered into three major genotypes. J. Med. Virol. 2006, 78, 517–522. [Google Scholar] [CrossRef]
- Quinlivan, M.; Hawrami, K.; Barrett-Muir, W.; Aaby, P.; Arvin, A.; Chow, V.T.; John, T.J.; Matondo, P.; Peiris, M.; Poulsen, A.; et al. The molecular epidemiology of varicella-zoster virus: Evidence for geographic segregation. J. Infect. Dis. 2002, 186, 888–894. [Google Scholar] [CrossRef]
- Rasmussen, L.; Geissler, A.; Cowan, C.; Chase, A.; Winters, M. The Genes Encoding the gCIII Complex of Human Cytomegalovirus Exist in Highly Diverse Combinations in Clinical Isolates. J. Virol. 2002, 76, 10841–10848. [Google Scholar] [CrossRef]
- Rasmussen, L.; Geissler, A.; Winters, M. Inter- and Intragenic Variations Complicate the Molecular Epidemiology of Human Cytomegalovirus. J. Infect. Dis. 2003, 187, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Rawlinson, W.D.; Farrell, H.E.; Barrell, B.G. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 1996, 70, 8833–8849. [Google Scholar] [CrossRef] [PubMed]
- Renzette, N.; Gibson, L.; Bhattacharjee, B.; Fisher, D.; Schleiss, M.R.; Jensen, J.D.; Kowalik, T.F. Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection. PLoS Genet. 2013, 9, e1003735. [Google Scholar] [CrossRef]
- Renzette, N.; Pokalyuk, C.; Gibson, L.; Bhattacharjee, B.; Schleiss, M.R.; Hamprecht, K.; Yamamoto, A.Y.; Mussi-Pinhata, M.M.; Britt, W.J.; Jensen, J.D. Limits and patterns of cytomegalovirus genomic diversity in humans. Proc. Natl. Acad. Sci. USA 2015, 112, E4120–E4128. [Google Scholar] [CrossRef]
- Retière, C.; Lesimple, B.; Lepelletier, D.; Bignon, J.-D.; Hallet, M.-M.; Imbert-Marcille, B.-M. Association of glycoprotein B and immediate early-1 genotypes with human leukocyte antigen alleles in renal transplant recipients with cytomegalovirus infection. Transplantation 2003, 75, 161. [Google Scholar] [CrossRef]
- Revello, M.G.; Gerna, G. Human cytomegalovirus tropism for endothelial/epithelial cells: Scientific background and clinical implications. Rev. Med. Virol. 2010, 20, 136–155. [Google Scholar] [CrossRef]
- Ross, S.A.; Arora, N.; Novak, Z.; Fowler, K.B.; Britt, W.J.; Boppana, S.B. Cytomegalovirus Reinfections in Healthy Seroimmune Women. J. Infect. Dis. 2010, 201, 386–389. [Google Scholar] [CrossRef]
- Ross, S.A.; Novak, Z.; Pati, S.; Patro, R.K.; Blumenthal, J.; Danthuluri, V.R.; Ahmed, A.; Michaels, M.G.; Sánchez, P.J.; Bernstein, D.I.; et al. Mixed Infection and Strain Diversity in Congenital Cytomegalovirus Infection. J. Infect. Dis. 2011, 204, 1003–1007. [Google Scholar] [CrossRef]
- Sakaue, S.; Gurajala, S.; Curtis, M.; Luo, Y.; Choi, W.; Ishigaki, K.; Kang, J.B.; Rumker, L.; Deutsch, A.J.; Schönherr, S.; et al. Tutorial: A statistical genetics guide to identifying HLA alleles driving complex disease. Nat. Protoc. 2023, 18, 2625–2641. [Google Scholar] [CrossRef]
- Sapuan, S.; Theodosiou, A.A.; Strang, B.L.; Heath, P.T.; Jones, C.E. A systematic review and meta-analysis of the prevalence of human cytomegalovirus shedding in seropositive pregnant women. Rev. Med. Virol. 2022, 32, e2399. [Google Scholar] [CrossRef]
- Sekulin, K.; Görzer, I.; Heiss-Czedik, D.; Puchhammer-Stöckl, E. Analysis of the variability of CMV strains in the RL11D domain of the RL11 multigene family. Virus Genes 2007, 35, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Shimamura, M.; Mach, M.; Britt, W.J. Human Cytomegalovirus Infection Elicits a Glycoprotein M (gM)/gN-Specific Virus-Neutralizing Antibody Response. J. Virol. 2006, 80, 4591–4600. [Google Scholar] [CrossRef] [PubMed]
- Sijmons, S.; Thys, K.; Mbong Ngwese, M.; Van Damme, E.; Dvorak, J.; Van Loock, M.; Li, G.; Tachezy, R.; Busson, L.; Aerssens, J.; et al. High-Throughput Analysis of Human Cytomegalovirus Genome Diversity Highlights the Widespread Occurrence of Gene-Disrupting Mutations and Pervasive Recombination. J. Virol. 2015, 89, 7673–7695. [Google Scholar] [CrossRef]
- Sijmons, S.; Van Ranst, M.; Maes, P. Genomic and Functional Characteristics of Human Cytomegalovirus Revealed by Next-Generation Sequencing. Viruses 2014, 6, 1049–1072. [Google Scholar] [CrossRef]
- Sinzger, C.; Hahn, G.; Digel, M.; Katona, R.; Sampaio, K.L.; Messerle, M.; Hengel, H.; Koszinowski, U.; Brune, W.; Adler, B. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 2008, 89, 359–368. [Google Scholar] [CrossRef]
- Stanton, R.J.; Baluchova, K.; Dargan, D.J.; Cunningham, C.; Sheehy, O.; Seirafian, S.; McSharry, B.P.; Neale, M.L.; Davies, J.A.; Tomasec, P.; et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Investig. 2010, 120, 3191–3208. [Google Scholar] [CrossRef]
- Stanton, R.; Westmoreland, D.; Fox, J.D.; Davison, A.J.; Wilkinson, G.W.G. Stability of human cytomegalovirus genotypes in persistently infected renal transplant recipients. J. Med. Virol. 2005, 75, 42–46. [Google Scholar] [CrossRef]
- Stern-Ginossar, N.; Weisburd, B.; Michalski, A.; Le, V.T.K.; Hein, M.Y.; Huang, S.-X.; Ma, M.; Shen, B.; Qian, S.-B.; Hengel, H.; et al. Decoding Human Cytomegalovirus. Science 2012, 338, 1088–1093. [Google Scholar] [CrossRef]
- Suárez, N.M.; Blyth, E.; Li, K.; Ganzenmueller, T.; Camiolo, S.; Avdic, S.; Withers, B.; Linnenweber-Held, S.; Gwinner, W.; Dhingra, A.; et al. Whole-Genome Approach to Assessing Human Cytomegalovirus Dynamics in Transplant Patients Undergoing Antiviral Therapy. Front. Cell. Infect. Microbiol. 2020, 10, 267. [Google Scholar] [CrossRef]
- Suárez, N.M.; Musonda, K.G.; Escriva, E.; Njenga, M.; Agbueze, A.; Camiolo, S.; Davison, A.J.; Gompels, U.A. Multiple-Strain Infections of Human Cytomegalovirus With High Genomic Diversity Are Common in Breast Milk From Human Immunodeficiency Virus–Infected Women in Zambia. J. Infect. Dis. 2019, 220, 792–801. [Google Scholar] [CrossRef]
- Suárez, N.M.; Wilkie, G.S.; Hage, E.; Camiolo, S.; Holton, M.; Hughes, J.; Maabar, M.; Vattipally, S.B.; Dhingra, A.; Gompels, U.A.; et al. Human Cytomegalovirus Genomes Sequenced Directly From Clinical Material: Variation, Multiple-Strain Infection, Recombination, and Gene Loss. J. Infect. Dis. 2019, 220, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Szpara, M.L.; Gatherer, D.; Ochoa, A.; Greenbaum, B.; Dolan, A.; Bowden, R.J.; Enquist, L.W.; Legendre, M.; Davison, A.J. Evolution and diversity in human herpes simplex virus genomes. J. Virol. 2014, 88, 1209–1227. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Kawata, M. A comprehensive test for negative frequency-dependent selection. Popul. Ecol. 2013, 55, 499–509. [Google Scholar] [CrossRef]
- Urban, M.; Klein, M.; Britt, W.J.; Haßfurther, E.; Mach, M. Glycoprotein H of human cytomegalovirus is a major antigen for the neutralizing humoral immune response. J. Gen. Virol. 1996, 77, 1537–1547. [Google Scholar] [CrossRef]
- Van Damme, E.; Van Loock, M. Functional annotation of human cytomegalovirus gene products: An update. Front. Microbiol. 2014, 5, 218. [Google Scholar] [CrossRef]
- Venturini, C.; Colston, J.M.; Charles, O.; Best, T.; Atkinson, C.; Forrest, C.; Williams, C.; Rao, K.; Worth, A.; Thorburn, D.; et al. Persistent low-level variants in a subset of HCMV genes are highly predictive of poor outcome in immunocompromised patients with cytomegalovirus infection. medRxiv 2022. [Google Scholar] [CrossRef]
- Venturini, C.; Pang, J.; Tamuri, A.U.; Roy, S.; Atkinson, C.; Griffiths, P.; Breuer, J.; Goldstein, R.A. Haplotype assignment of longitudinal viral deep-sequencing data using co-variation of variant frequencies. Virus Evol. 2022, 8, veac093. [Google Scholar] [CrossRef]
- Wada, K.; Mizuno, S.; Kato, K.; Kamiya, T.; Ozawa, K. Cytomegalovirus Glycoprotein B Sequence Variation among Japanese Bone Marrow Transplant Recipients. Intervirology 2008, 40, 215–219. [Google Scholar] [CrossRef]
- Walker, A.; Petheram, S.J.; Ballard, L.; Murph, J.R.; Demmler, G.J.; Bale, J.F. Characterization of Human Cytomegalovirus Strains by Analysis of Short Tandem Repeat Polymorphisms. J. Clin. Microbiol. 2001, 39, 2219–2226. [Google Scholar] [CrossRef]
- Wang, D.; Shenk, T. Human Cytomegalovirus UL131 Open Reading Frame Is Required for Epithelial Cell Tropism. J. Virol. 2005, 79, 10330–10338. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Valencia, S.M.; Pfeifer, S.P.; Jensen, J.D.; Kowalik, T.F.; Permar, S.R. Common Polymorphisms in the Glycoproteins of Human Cytomegalovirus and Associated Strain-Specific Immunity. Viruses 2021, 13, 1106. [Google Scholar] [CrossRef] [PubMed]
- Wegner, F.; Lassalle, F.; Depledge, D.P.; Balloux, F.; Breuer, J. Coevolution of Sites under Immune Selection Shapes Epstein–Barr Virus Population Structure. Mol. Biol. Evol. 2019, 36, 2512–2521. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, J.O.; Kosakovsky Pond, S.L. Purifying Selection Can Obscure the Ancient Age of Viral Lineages. Mol. Biol. Evol. 2011, 28, 3355–3365. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Prager, A.; Boos, S.; Resch, M.; Brizic, I.; Mach, M.; Wildner, S.; Scrivano, L.; Adler, B. Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog. 2017, 13, e1006281. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Koyano, S.; Inami, Y.; Yamamoto, Y.; Suzutani, T.; Mizuguchi, M.; Ushijima, H.; Kurane, I.; Inoue, N. Genetic linkage among human cytomegalovirus glycoprotein N (gN) and gO genes, with evidence for recombination from congenitally and post-natally infected Japanese infants. J. Gen. Virol. 2008, 89, 2275–2279. [Google Scholar] [CrossRef]
- Yan, H.; Koyano, S.; Inami, Y.; Yamamoto, Y.; Suzutani, T.; Mizuguchi, M.; Ushijima, H.; Kurane, I.; Inoue, N. Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch. Virol. 2008, 153, 667–674. [Google Scholar] [CrossRef]
- Zuhair, M.; Smit, G.S.A.; Wallis, G.; Jabbar, F.; Smith, C.; Devleesschauwer, B.; Griffiths, P. Estimation of the worldwide seroprevalence of cytomegalovirus: A systematic review and meta-analysis. Rev. Med. Virol. 2019, 29, e2034. [Google Scholar] [CrossRef]

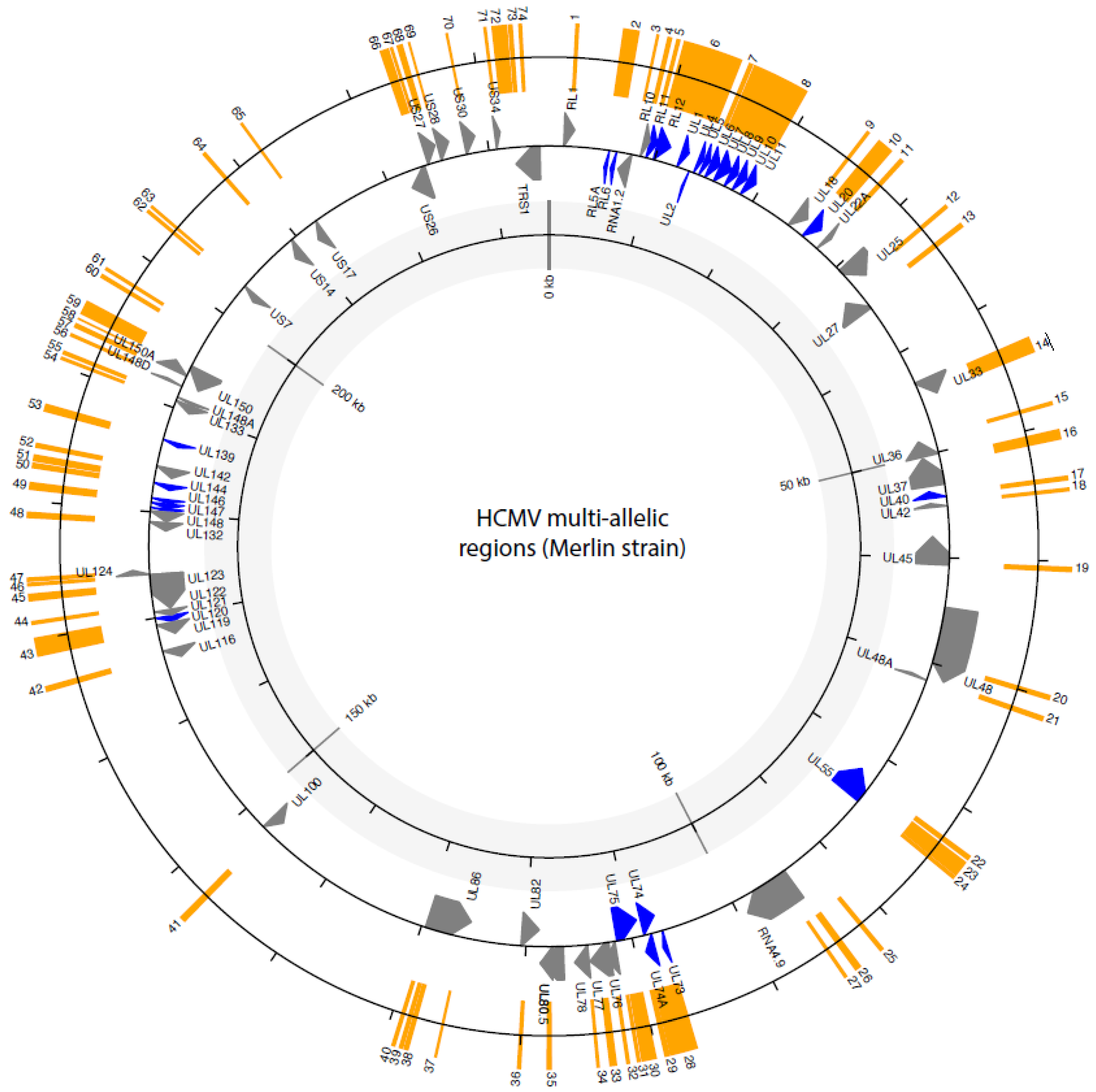
| Multi-Allelic Regions | LD | Genotypes Identified | Pseudogenes | Link Between HCMV Alleles and Function | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Genes | Start | End | N of Alleles | Geographic Allele Distribution | Previous Identified Genotypes | Most Variable Region | Hypervariable Genotypes Regions Suarez et al., 2019 | Geographic Genotypes Distribution | Phenotypes | Disease | ||
| 1 | RL1 | 1941 | 2121 | 2 | No | Yes | |||||||
| 2 | RL5A RL6 | 5387 | 6479 | 5 | Yes | Yes | RL5A: 6; RL6: 7 | Not described | Yes |
| |||
| 3 | RL9A | 7813 | 7914 | 2 | No | ||||||||
| 4 | RL10 | 8620 | 8868 | 3 | Yes | ||||||||
| 5 | RL11 | 9286 | 9479 | 2 | No | Yes | |||||||
| 6 | RL11 RL12 RL13 UL1 UL2 UL4 | 9840 | 14133 | 5 | No | Yes | UL1: 3; UL4: 4 (Sekulin, 2007) | RL12: 10 (+subtypes); RL13: 10 (+subtypes); UL1: 10 | UL1, RL13, RL12 | ||||
| 7 | UL5 UL6 | 14765 | 14993 | 2 | No | Yes | UL6: 4 (Sekulin, 2007) | ||||||
| 8 | UL10 UL11 UL6 UL7 UL8 UL9 | 15163 | 19324 | 4 | Yes | Yes | UL7: 3; UL10: 3 (Sekulin, 2007) | UL9: 9; UL11: 7 | UL9, UL11 | ||||
| 10 | UL20 | 25622 | 26757 | 3 | No | Yes | 7 | ||||||
| 17 | UL40 UL41A | 53875 | 54131 | 2 | No | Region encoding the HLA-E-binding peptide (residues 15–23 in AD169) (Heatley, 2013) | Yes |
| Not clear (Hartley 2013) | ||||
| 22 | UL55 | 82720 | 83003 | 2 | No | Yes | gB-5 genotypes (gB-1 to gB-5) (Wang, 2021) | Codons 26–70, gp55 cleavage site (codon 460) | All five genotypes have been detected in Asia, Europe, and North America; however, their distributions differ (Wang, 2022) |
| Studies show inconsistent associations between gB genotypes and CMV disease severity or clinical manifestations (Pati, 2013; Yan, 2008; Tarrago, 2003). cCMV Studies: Research indicates no consistent link between gB genotypes and symptoms, sensorineural hearing loss, or neurodevelopmental outcomes in congenital CMV cases (Pati, 2013; Arav-Boger, 2002; Bale, 2000). Some studies suggest specific genotypes like gB-3 may be more prevalent, but findings vary (Yan et al., 2008; Dong et al., 2023). Transmission and Clinical Outcomes: Studies across HIV and transplant patients show gB genotypes do not correlate with clinical outcomes, though specific genotypes may be associated with complications in transplant recipients (Tarrago, 2003; Torok-Storb, 1997; Dieamant, 2013). | ||
| 23 | UL55 | 83278 | 84403 | 3 | No | ||||||||
| 24 | UL55 | 84532 | 84716 | 3 | Yes | ||||||||
| 28 | UL73 | 107059 | 109022 | 7 | Yes | gN 4 genotypes gN-3 2 subtypes (Wang, 2021) | N terminal region | 4 (+subtypes) | Not described |
| Inconsistent findings (Arav-Boger, 2015) cCMV Disease Studies:
Transplant Recipients: No gN genotype was associated with a poorer outcome in solid organ transplant (SOT) recipients with CMV disease (Lisboa, 2012). | ||
| UL74 | 5 (gO-1 to gO-5) + subtypes (Wang, 2021) | N-terminal region (codons 1–98), codons 270–313 | 5 (+ subtypes) | Differences in g) genotypes distribution in Japanese children vs. European samples (Wang, 2021; Yan, 2008) | Deletions at the N-terminus (in the first 90 aa) (Rasmussen, 2002) |
| |||||||
| 29 | UL75 | 109129 | 109426 | 2 | No | Yes | 2 | N-terminal region (codons 1–37) | Not described |
| cCMV Disease Studies:
| ||
| 30 | UL75 | 110100 | 111111 | 2 | No | ||||||||
| 31 | UL75 | 111275 | 111445 | 2 | No | ||||||||
| 43 | UL119 UL120 UL121 | 168817 | 170109 | 4 | No | Yes | UL120: 4 (+subtypes) | ||||||
| 49 | UL146 UL147 | 180852 | 181323 | 8 | Yes | 14 (G1–G14) Bradley, 2008; Dolan, 2004) | UL146: 14 | Not described | Deleted in highly passaged lab strains (Cha, 1996) |
| cCMV Disease Studies: Inconclusive.
| ||
| 50 | UL144 | 182416 | 182725 | 3 | No | Yes | 3 (Arav-Boger, 2015) | No | Deleted in highly passaged lab strains (Cha, 1996) |
| cCMV Disease Studies: Controversial Findings (Arav-Boger, 2015).
| ||
| 53 | UL139 | 186573 | 187057 | 4 | No | 3-8 (Qi, 2006; Bradley, 2008) | N-terminal portion (Bradley, 2008) | 8 (+subtypes) | Not clear (Bradley 2008) | Deleted in highly passaged lab strains (Cha, 1996) | Shared sequence homology with human CD24 (signal transducer modulating B-cell activation responses). G1c contained a specific attachment site of prokaryotic membrane lipoprotein lipid (Qi, 2006). | ||
| 66 | US26 US27 | 223336 | 223914 | 2 | No | Yes | 5 | US27 | Functional beta-chemokine receptor | No association with cCMV disease (Pati, 2013; Arav Boger, 2002) | |||
| 67 | US27 | 224108 | 224224 | 3 | No | ||||||||
| 68 | US27 | 224607 | 224958 | 2 | No | ||||||||
| 69 | US27 US28 | 225456 | 225513 | 4 | No | ||||||||
| Multi-Allelic Regions | Previously Identified Variable Regions in gB |
|---|---|
| reg 24: codons 24–85 | codons 26–70 |
| reg 23: codons 128–503 | Codons 181–195; 311–317; gp55 cleavage site (codons 460). |
| reg 22: codons 595–689 | Not identified. |
| Haplotype | Example Strain | Previously Identified Genotypes |
|---|---|---|
| H1 | KY490079.1 | gB-4 |
| H2 | FJ527563.1 AD169 | gB-2 |
| H3 | KJ361956.1 | gB-4 |
| H4 | KY490069.1 | gB-2 |
| H5 | NC_006273.2 Merlin | gB-1 |
| H6 | FJ616285.1 Towne | gB-1 |
| H7 | KY490067.1 | Separate cluster—closer to gB-4 |
| H8 | GU179289.1 VR1814 | gB-3 |
| H9 | KY490088.1 | Separate cluster—closer to gB-4 |
| H10 | KJ361971.1 | gB-5 |
| Allele | Example Strain | Previously Identified Genotypes | Frequency in Europe | Frequency in America | Frequency in Africa |
|---|---|---|---|---|---|
| 1 | KY490061.1 | gN-3a | 20.5% | 9.1% | 23.3% |
| 2 | KY490065.1 | gN-3b | 8.4% | 27.3% | 3.3% |
| 3 | FJ616285.1 Towne | gN-4b | 11.6% | 18.2% | 66.7% |
| 4 | NC_006273.2 Merlin | gN-4c | 11.6% | 0 | 0 |
| 5 | KY490062.1 | gN-4a | 22.8% | 0 | 0 |
| 6 | FJ527563.1 | gN-1 | 13.5% | 9.1% | 6.7% |
| 7 | KJ361956.1 | gN-2 | 11.6% | 36.4% | 0 |
| Allele | Example Strain | Previously Identified gO Genotypes | Frequency in Europe | Frequency in America | Frequency in Africa |
|---|---|---|---|---|---|
| 1 | KY490061.1 | gO-1b | 20.5% | 9.1% | 23.3% |
| 2 | KY490065.1 | gO-2a | 8.4% | 27.3% | 3.3% |
| 3 | FJ616285.1 Towne | gO-4 | 11.6% | 18.2% | 66.7% |
| 4 | NC_006273.2 Merlin | gO-5 | 11.6% | 0 | 0 |
| 5 | KY490062.1 | gO-3 | 22.8% | 0 | 0 |
| 6 | FJ527563.1 AD169 | gO-1a | 13.5% | 9.1% | 6.7% |
| 7 | KJ361956.1 | gO-2b | 11.6% | 36.4% | 0 |
| Multi-Allelic Regions | Previously Identified Most Variable Regions in gH |
|---|---|
| reg 31: codons 3–60 | codons 1–37 |
| reg 30: codons 114–451 | Not identified. |
| reg 29: codons 676–742 | Not identified. |
| Allele | Strain | Genotype | Frequency in Europe | Frequency in America | Frequency in Africa |
|---|---|---|---|---|---|
| H1 | KY490061.1 | gH-1 | 26.05% | 0 | 30% |
| H2 | NC_006273.2 Merlin/FJ616285.1 Towne | gH-2 | 27.3% | 40% | 40% |
| H3 | FJ527563.1 AD169 | gH-1 | 20% | 9.1% | 13% |
| H4 | JX512206.1 | hybrid | 1.86% | 0 | 13% |
| H5 | KJ361946.1 | gH-2 | 10.70% | 36.4% | 0 |
| H6 | KP745640.1 | hybrid | 1.40% | 27.3% | 0 |
| Allele | Strain | Genotype | Frequency in Europe | Frequency in America | Frequency in Africa |
|---|---|---|---|---|---|
| A1 | NC_006273.2 Merlin | Group 1 | 39.5% | 9% | 60% |
| A2 | FJ616285.1 Towne/MF084224.1 | Group 3 | 45.6% | 55% | 23.3% |
| A3 | KY490064.1 | Group 2 | 14.9% | 36% | 16.7% |
| Allele | Strain | Genotype | Frequency in Europe | Frequency in America | Frequency in Africa |
|---|---|---|---|---|---|
| A1 | KY490068.1 | 13 | 13.49% | 27.3% | 3.3% |
| A2 | KY490084.1 | 7 | 2.33% | 18.2% | 3.3% |
| A3 | MK290742.1 | 12 | 17.67% | 9.1% | 20% |
| A4 | KY490088.1 | 9 | 12.09% | 0 | 50% |
| A5 | MK290743.1 | 5 | 13.95% | 9.1% | 13.3% |
| A6 | MF084224.1 | 8 | 16.28% | 9.1% | 0 |
| A7 | NC_006273.2 Merlin | 2 | 13.02% | 18.2% | 0 |
| A8 | KY490067.1 | 11 | 11.16% | 9.1% | 10% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venturini, C.; Breuer, J. Cytomegalovirus Genetic Diversity and Evolution: Insights into Genotypes and Their Role in Viral Pathogenesis. Pathogens 2025, 14, 50. https://doi.org/10.3390/pathogens14010050
Venturini C, Breuer J. Cytomegalovirus Genetic Diversity and Evolution: Insights into Genotypes and Their Role in Viral Pathogenesis. Pathogens. 2025; 14(1):50. https://doi.org/10.3390/pathogens14010050
Chicago/Turabian StyleVenturini, Cristina, and Judith Breuer. 2025. "Cytomegalovirus Genetic Diversity and Evolution: Insights into Genotypes and Their Role in Viral Pathogenesis" Pathogens 14, no. 1: 50. https://doi.org/10.3390/pathogens14010050
APA StyleVenturini, C., & Breuer, J. (2025). Cytomegalovirus Genetic Diversity and Evolution: Insights into Genotypes and Their Role in Viral Pathogenesis. Pathogens, 14(1), 50. https://doi.org/10.3390/pathogens14010050






