Genomic Insight into Vibrio Isolates from Fresh Raw Mussels and Ready-to-Eat Stuffed Mussels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Isolation
2.3. MALDI-TOF MS Identification
2.4. Genome Sequencing, Assembly, and Identification
2.5. Genome Analysis
3. Results
3.1. Sequence Analysis
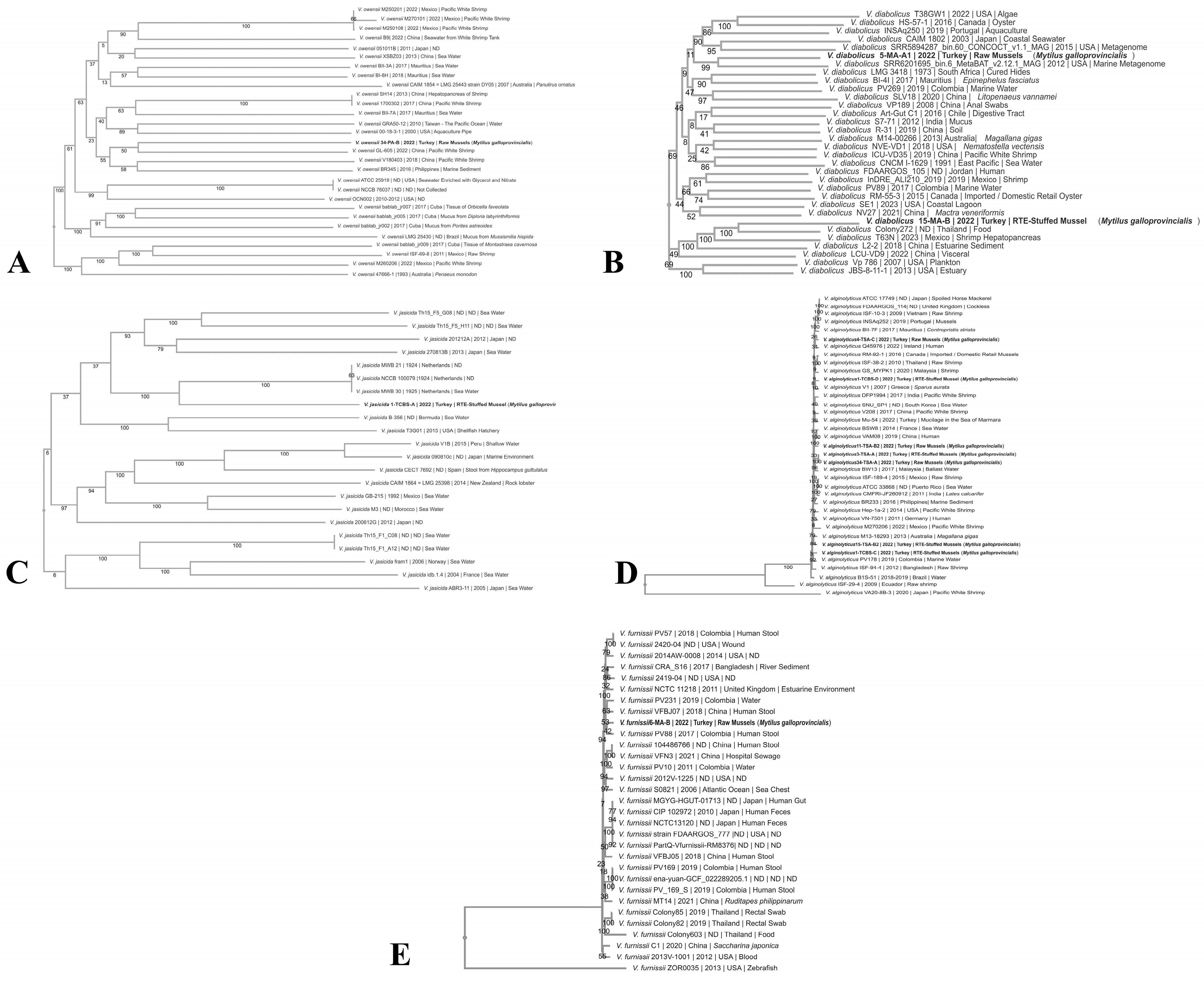
3.2. Phylogenomics
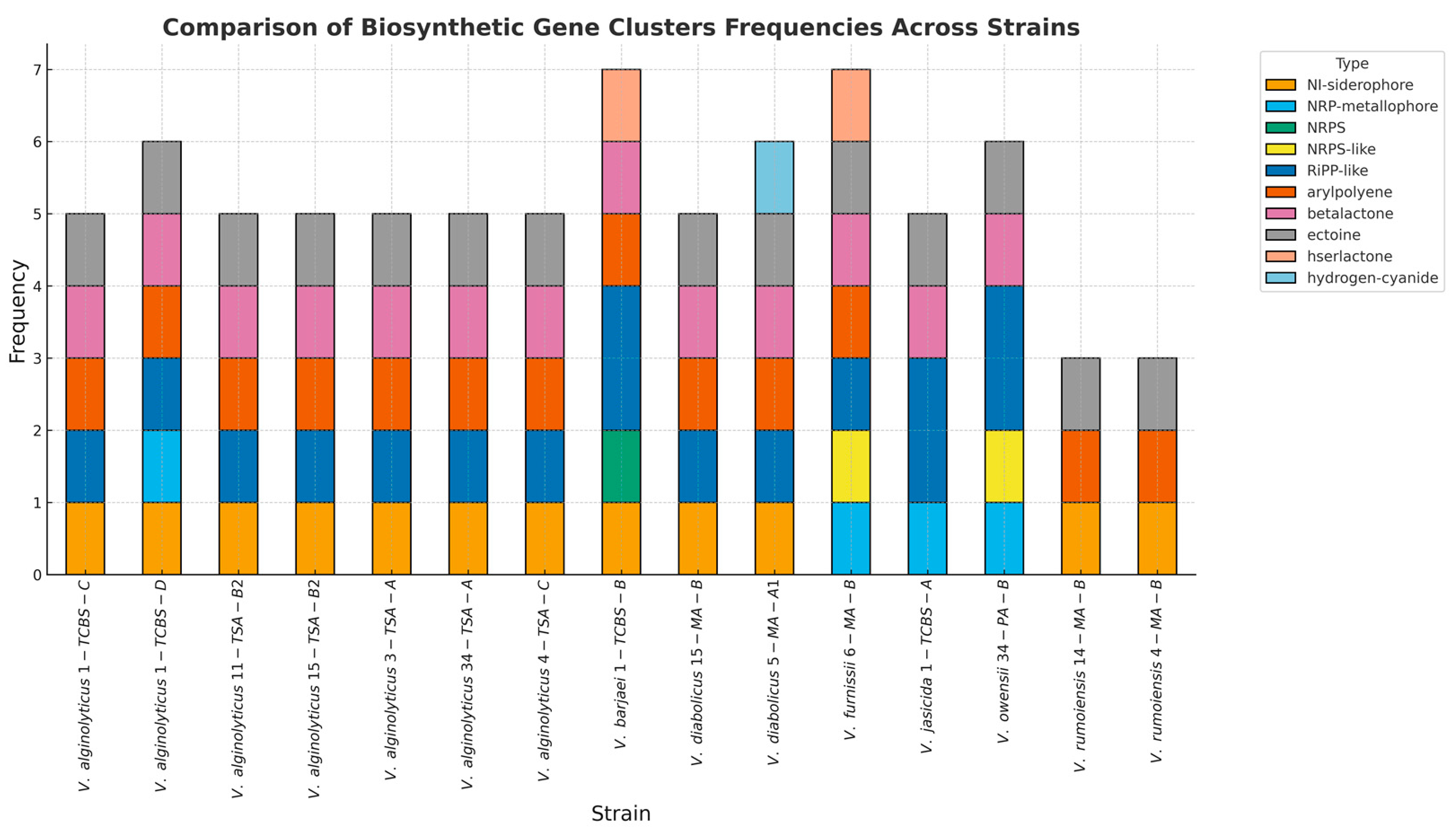
3.3. Biosynthetic Gene Clusters (BGCs) and Prophage
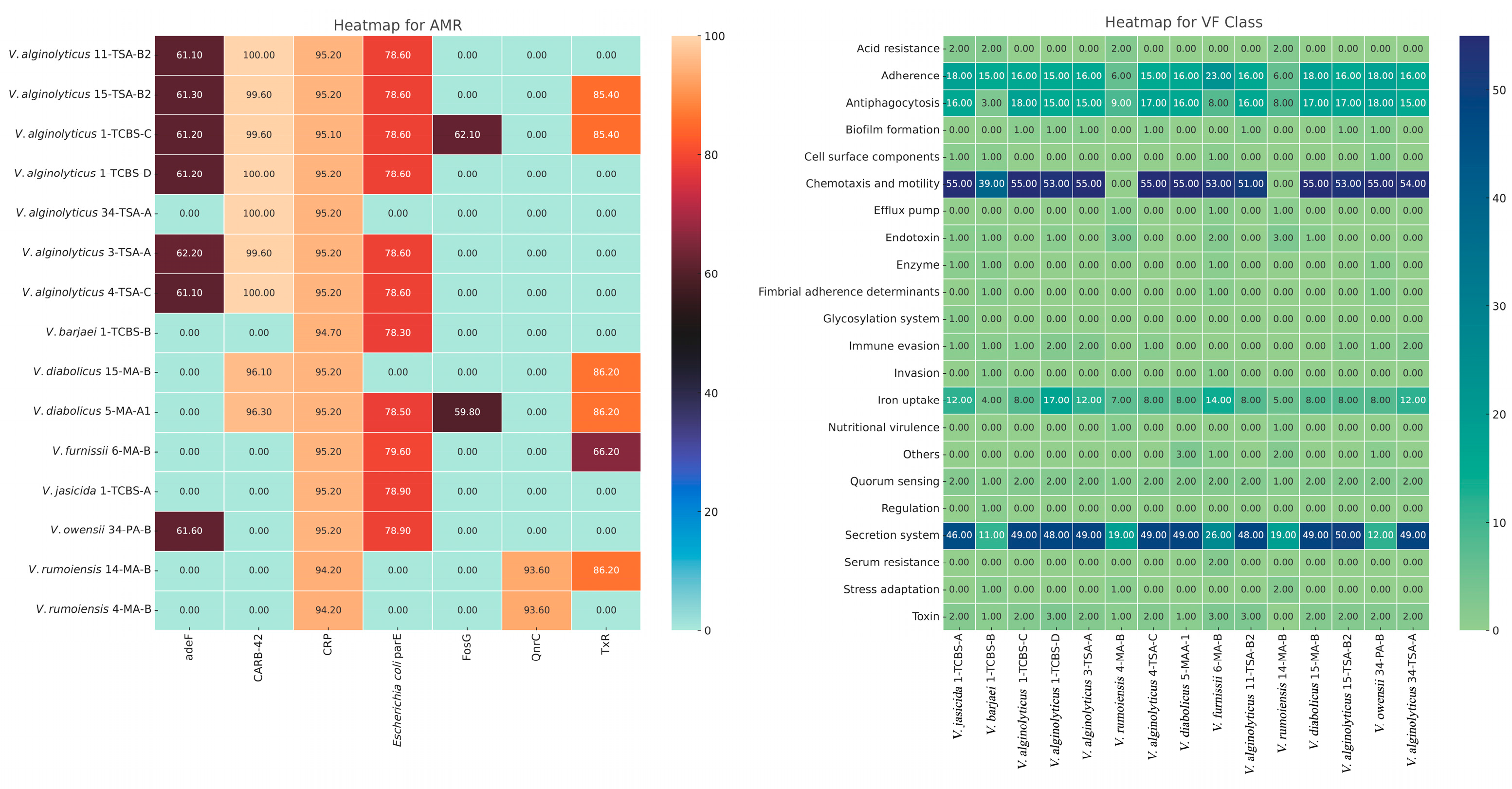
3.4. Antimicrobial Resistance (AMR) and Virulence Factor (VF) Genes
3.5. Potential Human Pathogenicity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ayvaz, Z. Geleneksel bir ürün olarak “Midye Dolma” ve gelecek önerileri. Ziraat Mühendisliği 2018, 366, 21–27. [Google Scholar] [CrossRef]
- Turkish Statistical Institute (TURKSTAT). Annual Statistical Report 2023. Available online: https://data.tuik.gov.tr/Bulten/Index?p=Su-Urunleri-2023-53702 (accessed on 16 December 2024).
- World Bank. Turkey Export Data for Product 030731 in 2023. Available online: https://wits.worldbank.org/trade/comtrade/en/country/TUR/year/2023/tradeflow/Exports/partner/ALL/product/030731 (accessed on 16 December 2024).
- Kahraman, Y.D.; Berik, N. Phenotypic and genotypic antibiotic resistance of Staphylococcus warneri and Staphylococcus pasteuri isolated from stuffed mussels. Aquat. Sci. Eng. 2024, 39, 172–178. [Google Scholar] [CrossRef]
- Kisla, D.; Uzgun, Y. Microbiological evaluation of stuffed mussels. J. Food Prot. 2008, 71, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Non-cholera Vibrios: The microbial barometer of climate change. Trends Microbiol. 2017, 25, 76–84. [Google Scholar] [CrossRef]
- Destoumieux-Garzón, D.; Canesi, L.; Oyanedel, D.; Travers, M.-A.; Charrière, G.M.; Pruzzo, C.; Vezzulli, L. Vibrio–bivalve interactions in health and disease. Environ. Microbiol. 2020, 22, 4323–4341. [Google Scholar] [CrossRef] [PubMed]
- Martins, V.G.P.; Nascimento, J.D.S.; Martins, F.M.D.S.; Vigoder, H.C. Vibriosis and its impact on microbiological food safety. Food Sci. Technol. 2022, 42, 1–6. [Google Scholar] [CrossRef]
- Sampaio, A.; Silva, V.; Poeta, P.; Aonofriesei, F. Vibrio spp.: Life strategies, ecology, and risks in a changing environment. Diversity 2022, 14, 97. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA). Vibrio Bacteria in Seafood: Increased Risk Due to Climate Change and Antimicrobial Resistance. EFSA. 2024. Available online: https://www.efsa.europa.eu/en/news/vibrio-bacteria-seafood-increased-risk-due-climate-change-and-antimicrobial-resistance (accessed on 16 October 2024).
- Derber, C.; Coudron, P.; Tarr, C.; Gladney, L.; Turnsek, M.; Shankaran, S.; Wong, E. Vibrio furnissii: An unusual cause of bacteremia and skin lesions after ingestion of seafood. J. Clin. Microbiol. 2011, 49, 2348–2349. [Google Scholar] [CrossRef]
- Zhou, K.; Tian, K.Y.; Liu, X.Q.; Liu, W.; Zhang, X.Y.; Liu, J.Y.; Sun, F. Characteristic and otopathogenic analysis of a Vibrio alginolyticus strain responsible for chronic otitis externa in China. Front. Microbiol. 2021, 12, 750642. [Google Scholar] [CrossRef]
- Yumoto, I.; Iwata, H.; Sawabe, T.; Ueno, K.; Ichise, N.; Matsuyama, H.; Okuyama, H.; Kawasaki, K. Characterization of a facultatively psychrophilic bacterium, Vibrio rumoiensis sp. nov., that exhibits high catalase activity. Appl. Environ. Microbiol. 1999, 65, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Dubert, J.; Balboa, S.; Regueira, M.; González-Castillo, A.; Gómez-Gil, B.; Romalde, J.L. Vibrio barjaei sp. nov., a new species of the Mediterranei clade isolated in a shellfish hatchery. Syst. Appl. Microbiol. 2016, 39, 553–556. [Google Scholar] [CrossRef]
- Liu, L.; Xiao, J.; Zhang, M.; Zhu, W.; Xia, X.; Dai, X.; Pan, Y.; Yan, S.; Wang, Y. A Vibrio owensii strain as the causative agent of AHPND in cultured shrimp, Litopenaeus vannamei. J. Invertebr. Pathol. 2018, 153, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; He, X.; Austin, B. Vibrio harveyi: A serious pathogen of fish and invertebrates in mariculture. Mar. Life Sci. Technol. 2020, 2, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Liu, X.; Wu, C.; Zhang, Y.; Fan, K.; Zhang, X.; Wei, Y. Isolation, identification and pathogenesis study of Vibrio diabolicus. Aquaculture 2021, 533, 736043. [Google Scholar] [CrossRef]
- Deng, Y.; Xu, H.; Su, Y.; Liu, S.; Xu, L.; Guo, Z.; Wu, J.; Cheng, C.; Feng, J. Horizontal gene transfer contributes to virulence and antibiotic resistance of Vibrio harveyi 345 based on complete genome sequence analysis. BMC Genomics 2019, 20, 761. [Google Scholar] [CrossRef] [PubMed]
- Goulden, E.F.; Hall, M.R.; Bourne, D.G.; Pereg, L.L.; Høj, L. Pathogenicity and infection cycle of Vibrio owensii in larviculture of the Ornate Spiny Lobster (Panulirus ornatus). Appl. Environ. Microbiol. 2012, 78, 2841–2849. [Google Scholar] [CrossRef]
- Harrison, J.; Nelson, K.; Morcrette, H.; Morcrette, C.; Preston, J.; Helmer, L.; Titball, R.W.; Butler, C.S.; Wagley, S. The increased prevalence of Vibrio species and the first reporting of Vibrio jasicida and Vibrio rotiferianus at UK shellfish sites. Water Res. 2022, 211, 117942. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO); World Health Organization (WHO). Available online: https://openknowledge.fao.org/server/api/core/bitstreams/2b4234e8-a0e1-45b6-8574-e1560e6405cf/content (accessed on 17 October 2024).
- Mititelu, M.; Neacșu, S.M.; Oprea, E.; Dumitrescu, D.E.; Nedelescu, M.; Drăgănescu, D.; Nicolescu, T.O.; Roșca, A.C.; Ghica, M. Black Sea mussels qualitative and quantitative chemical analysis: Nutritional benefits and possible risks through consumption. Nutrients 2022, 14, 964. [Google Scholar] [CrossRef]
- Bakshi, B.; Bouchard, R.W., Jr.; Dietz, R.; Hornbach, D.; Monson, P.; Sietman, B.; Wasley, D. Freshwater mussels, ecosystem services, and clean water regulation in Minnesota: Formulating an effective conservation strategy. Water 2023, 15, 2560. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Available online: https://www.cdc.gov/vibrio/about/index.html (accessed on 17 October 2024).
- Kumarage, P.M.; De Silva, L.A.D.S.; Heo, G.J. Aquatic environments: A potential source of antimicrobial-resistant Vibrio spp. J. Appl. Microbiol. 2022, 133, 2267–2279. [Google Scholar] [CrossRef] [PubMed]
- Stephen, J.; Lekshmi, M.; Ammini, P.; Kumar, S.H.; Varela, M.F. Membrane efflux pumps of pathogenic Vibrio species: Role in antimicrobial resistance and virulence. Microorganisms 2022, 10, 382. [Google Scholar] [CrossRef] [PubMed]
- Yibar, A.; Saticioglu, I.B.; Ajmi, N.; Duman, M. Molecular characterization and antibacterial resistance determination of Escherichia coli isolated from fresh raw mussels and ready-to-eat stuffed mussels: A major public health concern. Pathogens 2024, 13, 532. [Google Scholar] [CrossRef] [PubMed]
- Vural, P.; Yildiz, H.; Acarli, S. Growth and survival performances of Mediterranean mussel (Mytilus galloprovincialis, Lamarck, 1819) on different depths in Cardak lagoon, Dardanelles. Mar. Sci. Technol. Bull. 2015, 4, 7–12. [Google Scholar]
- Pretto, T. Vibrio harveyi group. In Diagnostic Manual for the Main Pathogens in European Seabass and Gilthead Seabream Aquaculture; Zrncic, S., Ed.; International Centre for Advanced Mediterranean Agronomic Studies (CIHEAM): Zaragoza, Spain, 2020; pp. 75–82. [Google Scholar]
- Vázquez-Rosas-Landa, M.; Ponce-Soto, G.Y.; Aguirre-Liguori, J.A.; Thakur, S.; Scheinvar, E.; Barrera-Redondo, J.; Ibarra-Laclette, E.; Guttman, D.S.; Eguiarte, L.E.; Souza, V. Population genomics of Vibrionaceae isolated from an endangered oasis reveals local adaptation after an environmental perturbation. BMC Genom. 2020, 21, 418. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Xia, F. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD, and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. AntiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
- Bertelli, C.; Gray, K.L.; Woods, N.; Lim, A.C.; Tilley, K.E.; Winsor, G.L.; Brinkman, F.S.L. Enabling genomic island prediction and comparison in multiple genomes to investigate bacterial evolution and outbreaks. Microb. Genom. 2022, 8, 000818. [Google Scholar] [CrossRef] [PubMed]
- Kary, S.C.; Yoneda, J.R.K.; Olshefsky, S.C.; Stewart, L.A.; West, S.B.; Cameron, A.D.S. The global regulatory cyclic AMP receptor protein (CRP) controls multifactorial fluoroquinolone susceptibility in Salmonella enterica Serovar Typhimurium. Antimicrob. Agents Chemother. 2017, 61, e01666-17. [Google Scholar] [CrossRef]
- Shmakov, S.A.; Utkina, I.; Wolf, Y.I.; Makarova, K.S.; Severinov, K.V.; Koonin, E.V. CRISPR Arrays Away from Cas Genes. CRISPR J. 2020, 3, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Beshiru, A.; Igbinosa, E.O. Surveillance of Vibrio parahaemolyticus pathogens recovered from ready-to-eat foods. Sci. Rep. 2023, 13, 4186. [Google Scholar] [CrossRef] [PubMed]
- Ashrafudoulla, M.; Na, K.W.; Hossain, M.I.; Mizan, M.F.R.; Nahar, S.; Toushik, S.H.; Roy, P.K.; Park, S.H.; Ha, S. Molecular and pathogenic characterization of Vibrio parahaemolyticus isolated from seafood. Mar. Pollut. Bull. 2021, 172, 112927. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, O.M.; Amin, H.F. Detection and survival of Vibrio species in shrimp (Penaeus indicus) and mussel (Mytilus galloprovincialis) at landing and after processing at seafood markets in Suez, Egypt. J. Food Dairy Sci. 2018, 9, 411–417. [Google Scholar] [CrossRef]
- Tang, J.-Y.-H.; Farhana Sakinah, M.R.; Nakaguchi, Y.; Nishibuchi, M.; Chai, L.-C.; New, C.Y.; Son, R. Detection of Vibrio cholerae in street food (satar and otak-otak) by Loop-Mediated Isothermal Amplification (LAMP), multiplex Polymerase Chain Reaction (mPCR) and plating methods. Food Res. 2018, 2, 447–452. [Google Scholar] [CrossRef]
- Nuñal, S.N.; Jane M Monaya, K.; Rose T Mueda, C.; Mae Santander-De Leon, S. Microbiological quality of oysters and mussels along its market supply chain. J. Food Prot. 2023, 86, 100063. [Google Scholar] [CrossRef] [PubMed]
- Preeprem, S.; Aksonkird, T.; Nuidate, T.; Hajimasalaeh, W.; Hajiwangoh, Z.; Mittraparp-arthorn, P. Characterization and genetic relationships of Vibrio spp. isolated from seafood in retail markets, Yala, Thailand. Trends Sci. 2023, 20, 5962. [Google Scholar] [CrossRef]
- Kering, K.; Wang, Y.; Mbae, C.; Mugo, M.; Ongadi, B.; Odityo, G.; Muturi, P.; Yakubu, H.; Liu, P.; Durry, S.; et al. Pathways of exposure to Vibrio cholerae in an urban informal settlement in Nairobi, Kenya. PLOS Glob. Public Health 2024, 4, e0002880. [Google Scholar] [CrossRef]
- Zohra, T.; Ikram, A.; Salman, M.; Amir, A.; Saeed, A.; Ashraf, Z.; Ahad, A. Wastewater-based environmental surveillance of toxigenic Vibrio cholerae in Pakistan. PLoS ONE 2021, 16, e0257414. [Google Scholar] [CrossRef] [PubMed]
- Galbraith, P.S.; Larouche, P.; Chasse, J.; Petrie, B. Sea-surface temperature in relation to air temperature in the Gulf of St. Lawrence: Interdecadal variability and long term trends. Deep-Sea Res. Part II Top. Stud. Oceanogr. 2012, 77–80, 10–20. [Google Scholar] [CrossRef]
- Feng, X.; Haines, K.; de Boisseson, E. Coupling of surface air and sea surface temperatures in the CERA-20C re-analysis. Q. J. R. Meteorol. Soc. 2018, 144, 195–207. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Trinanes, J.A.; Taylor, N.G.; Hartnell, R.; Siitonen, A. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 2013, 3, 73–77. [Google Scholar] [CrossRef]
- Gu, D.; Guo, M.; Yang, M.; Zhang, Y.; Zhou, X.; Wang, Q. A ςe-mediated temperature gauge controls a switch from luxr-mediated virulence gene expression to thermal stress adaptation in Vibrio alginolyticus. PLoS Pathog. 2016, 12, e1005645. [Google Scholar] [CrossRef]
- Sheikh, H.I.; Najiah, M.; Fadhlina, A.; Laith, A.A.; Nor, M.M.; Jalal, K.C.A.; Kasan, N.A. Temperature upshift mostly but not always enhances the growth of Vibrio species: A systematic review. Front. Mar. Sci. 2022, 9, 959830. [Google Scholar] [CrossRef]
- Abd El Tawab, A.; Ibrahim, A.M.; Sittien, A. Phenotypic and genotypic characterization of Vibrio species isolated from marine fishes. Benha Vet. Med. J. 2018, 34, 79–93. [Google Scholar] [CrossRef]
- Mahmoud, S.; El-Bouhy, Z.; Hassanin, M.; Fadel, A.H. Vibrio alginolyticus and Photobacterium damselae subsp. damselae: Prevalence, histopathology and treatment in sea bass Dicentrarchus labrax. J. Pharm. Chem. Biol. Sci. 2018, 5, 354–364. [Google Scholar]
- Elgendy, M.; Soliman, W.; Hassan, H.; Kenawy, A.; Liala, A.M. Effect of abrupt environmental deterioration on the eruption of vibriosis in mari-cultured shrimp, Penaeus indicus, in Egypt. J. Fish Aquat. Sci. 2015, 10, 146–158. [Google Scholar] [CrossRef]
- Ismail, E.T.; El-Son, M.A.M.; El-Gohary, F.A.; Zahran, E. Prevalence, genetic diversity, and antimicrobial susceptibility of Vibrio spp. infected gilthead sea breams from coastal farms at Damietta, Egypt. BMC Vet. Res. 2024, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.G. Metabolic and molecular responses of fish to hypoxia. In Fish Physiology; Elsevier: Amsterdam, The Netherlands, 2009; Volume 27, pp. 443–485. [Google Scholar]
- Huang, Y.; Du, P.; Zhao, M.; Liu, W.; Du, Y.; Diao, B.; Li, J.; Kan, B.; Liang, W. Functional characterization and conditional regulation of the type VI secretion system in Vibrio fluvialis. Front. Microbiol. 2017, 8, 528. [Google Scholar] [CrossRef] [PubMed]
- Brauge, T.; Mougin, J.; Ells, T.; Midelet, G. Sources and contamination routes of seafood with human pathogenic Vibrio spp.: A farm-to-fork approach. Compreh. Rev. Food Sci. Food Saf. 2023, 23, e13283. [Google Scholar] [CrossRef] [PubMed]
- Al-Garadi, M.A.; Aziz, R.N.; Almashhadany, D.A.; Al Qabili, D.M.A.; Abdullah Aljoborey, A.D. Validity of cold storage and heat treatment on the deactivation of Vibrio parahaemolyticus isolated from fish meat markets. Ital. J. Food Saf. 2024, 13, 11516. [Google Scholar] [CrossRef] [PubMed]
- European Commission (EC). Opinion of the Scientific Committee on Veterinary Measures Relating to Public Health on Vibrio vulnificus and Vibrio parahaemolyticus (in Raw and Undercooked Seafood). European Commission. 2001. Available online: https://food.ec.europa.eu/system/files/2020-12/sci-com_scv_out45_en.pdf (accessed on 19 October 2024).
- Hara-Kudo, Y.; Kumagai, S. Impact of seafood regulations for Vibrio parahaemolyticus infection and verification by analyses of seafood contamination and infection. Epidemiol. Infect. 2014, 142, 2237–2247. [Google Scholar] [CrossRef] [PubMed]
- NSW Food Authority. Potentially Hazardous Foods—Foods that Require Temperature Control for Safety. NSW Government. 2023. Available online: https://www.foodauthority.nsw.gov.au/sites/default/files/_Documents/industry/potentially_hazardous_foods.pdf (accessed on 19 October 2024).
- Lin, H.; Yu, M.; Wang, X.; Zhang, X.H. Comparative genomic analysis reveals the evolution and environmental adaptation strategies of vibrios. BMC Genom. 2018, 19, 135. [Google Scholar] [CrossRef] [PubMed]
- Schwibbert, K.; Marin-Sanguino, A.; Bagyan, I.; Heidrich, G.; Lentzen, G.; Seitz, H.; Rampp, M.; Schuster, S.C.; Klenk, H.P.; Pfeiffer, F.; et al. A blueprint of ectoine metabolism from the genome of the industrial producer Halomonas elongata DSM 2581 T. Environ. Microbiol. 2011, 13, 1973–1994. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.; Osborn, L.J.; Markley, R.L.; McManus, E.A.; Kadam, A.; Schultz, K.B.; Nagajothi, N.; Ahern, P.P.; Brown, J.M.; Claesen, J. Identification of essential genes for Escherichia coli aryl polyene biosynthesis and function in biofilm formation. NPJ Biofilms Microbiomes 2021, 7, 56. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Choi, S.; Jang, A.; Son, K.; Kim, Y. Structural comparison of Acinetobacter baumannii β-ketoacyl-acyl carrier protein reductases in fatty acid and aryl polyene biosynthesis. Sci. Rep. 2021, 11, 7945. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Bacterial siderophores: Classification, biosynthesis, perspectives of use in agriculture. Plants 2022, 11, 3065. [Google Scholar] [CrossRef] [PubMed]
- Džunková, M.; La Clair, J.J.; Tyml, T.; Doud, D.; Schulz, F.; Piquer-Esteban, S.; Sanchis, D.P.; Osborn, A.; Robinson, D.; Louie, K.B.; et al. Synthase-selected sorting approach identifies a beta-lactone synthase in a nudibranch symbiotic bacterium. Microbiome 2023, 11, 130. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Rajput, V.D.; Prazdnova, E.V.; Gurnani, M.; Bhardwaj, P.; Sharma, S.; Sushkova, S.; Mandzhieva, S.S.; Minkina, T.; Sudan, J.; et al. Nature’s antimicrobial arsenal: Non-ribosomal peptides from PGPB for plant pathogen biocontrol. Fermentation 2023, 9, 597. [Google Scholar] [CrossRef]
- Yoo, Y.; Lee, H.; Lee, J.; Khim, J.S.; Kim, J. Insights into saline adaptation strategies through a novel halophilic bacterium isolated from solar saltern of Yellow Sea. Front. Mar. Sci. 2023, 10, 1229444. [Google Scholar] [CrossRef]
- Cai, H.; Yu, J.; Li, Q.; Zhang, Y.; Huang, L. Research Progress on Virulence Factors of Vibrio alginolyticus: A Key Pathogenic Bacteria of Sepsis; IntechOpen: London, UK, 2024. [Google Scholar] [CrossRef]
- Lee, W.C.; Choi, S.; Jang, A.; Yeon, J.; Hwang, E.; Kim, Y. Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis. Sci. Rep. 2021, 11, 16340. [Google Scholar] [CrossRef] [PubMed]
- Zane, H.K.; Naka, H.; Rosconi, F.; Sandy, M.; Haygood, M.G.; Butler, A. Biosynthesis of amphi-enterobactin siderophores by Vibrio harveyi BAA-1116: Identification of a bifunctional nonribosomal peptide synthetase condensation domain. J. Am. Chem. Soc. 2014, 136, 5615–5618. [Google Scholar] [CrossRef] [PubMed]
- Cunha-Ferreira, I.C.; Vizzotto, C.S.; Frederico, T.D.; Peixoto, J.; Carvalho, L.S.; Tótola, M.R.; Krüger, R.H. Impact of Paenibacillus elgii supernatant on screening bacterial strains with potential for biotechnological applications. Eng. Microbiol. 2024, 4, 100163. [Google Scholar] [CrossRef]
- Khalid, S.J.; Ain, Q.; Khan, S.J.; Jalil, A.; Siddiqui, M.F.; Ahmad, T.; Badshah, M.; Adnan, F. Targeting Acyl Homoserine Lactones (AHLs) by the quorum quenching bacterial strains to control biofilm formation in Pseudomonas aeruginosa. Saudi J. Biol. Sci. 2022, 29, 1673–1682. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, P.E.; Nielsen, T.K.; Riber, L.; Lading, H.H.; Milena, L.; Kot, W.; Raaijmakers, J.M.; Hansen, L.H. Widespread and largely unknown prophage activity, diversity, and function in two genera of wheat phyllosphere bacteria. ISME J. 2023, 17, 2415–2425. [Google Scholar] [CrossRef]
- Sorée, M. Towards a Better Overview of Virulence and Risk Management of Vibrio parahaemolyticus, a Marine Bacterium Potentially Pathogenic for Humans. Ph.D. Thesis, Université de Bretagne Occidentale, Brest, France, 2022. [Google Scholar]
- Neumann, B.; Lippmann, N.; Wendt, S.; Karlas, T.; Lübbert, C.; Werner, G.; Pfeifer, Y.; Schuster, C.F. Recurrent bacteremia with a hypermucoviscous Escherichia coli isolated from a patient with perihilar cholangiocarcinoma: Insights from a comprehensive genome-based analysis. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 28. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Piddock, L.J. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 2003, 51, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef]
- Piddock, L.J. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef]
- Schaenzer, A.J.; Wright, G.D. Antibiotic resistance by enzymatic modification of antibiotic targets. Trends Mol. Med. 2020, 26, 768–782. [Google Scholar] [CrossRef]
- De Pascale, G.; Wright, G.D. Antibiotic resistance by enzyme inactivation: From mechanisms to solutions. Chembiochem 2010, 11, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Karpov, D.S.; Kazakova, E.M.; Kovalev, M.A.; Shumkov, M.S.; Kusainova, T.; Tarasova, I.A.; Osipova, P.J.; Poddubko, S.V.; Mitkevich, V.A.; Kuznetsova, M.V.; et al. Determinants of antibiotic resistance and virulence factors in the genome of Escherichia coli APEC 36 strain isolated from a broiler chicken with generalized colibacillosis. Antibiotics 2024, 13, 945. [Google Scholar] [CrossRef] [PubMed]
- Hirshfeld, B.; Lavelle, K.; Lee, K.Y.; Atwill, E.R.; Kiang, D.; Bolkenov, B.; Gaa, M.; Li, Z.; Yu, A.; Li, X.; et al. Prevalence and antimicrobial resistance profiles of Vibrio spp. and Enterococcus spp. in retail shrimp in Northern California. Front. Microbiol. 2023, 14, 1192769. [Google Scholar] [CrossRef]
- Yasir, M.; Ullah, R.; Bibi, F.; Khan, S.B.; Al-Sofyani, A.A.; Stingl, U.; Azhar, E.I. Draft genome sequence of a multidrug-resistant emerging pathogenic isolate of Vibrio alginolyticus from the Red Sea. New Microbes New Infect. 2020, 38, 100804. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.M.; Mercoulia, K.; Valcanis, M.; Gorrie, C.L.; Sherry, N.L.; Howden, B.P. Hidden Resistances: How routine whole-genome sequencing uncovered an otherwise undetected blandm-1 gene in Vibrio alginolyticus from imported seafood. Microbiol. Spectrum 2023, 11, e0417622. [Google Scholar] [CrossRef]
- Rozman, U.; Pušnik, M.; Kmetec, S.; Duh, D.; Šostar Turk, S. Reduced susceptibility and increased resistance of bacteria against disinfectants: A systematic review. Microorganisms 2021, 9, 2550. [Google Scholar] [CrossRef]
- Ferri, G.; Olivieri, V.; Olivastri, A.; Pennisi, L.; Vergara, A. Multidrug resistant Vibrio spp. identified from mussels farmed for human consumption in Central Italy. J. Appl. Microbiol. 2024, 135, lxae098. [Google Scholar] [CrossRef]
- Abdelaziz Gobarah, D.E.; Helmy, S.M.; Mahfouz, N.B.; Fahmy, H.A.; Abou Zeid, M.A.E.H.M. Virulence genes and antibiotic resistance profile of Vibrio species isolated from fish in Egypt. Vet. Res. Forum 2022, 13, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Xu, L.; Chen, H.; Liu, S.; Guo, Z.; Cheng, C.; Ma, H.; Feng, J. Prevalence, virulence genes, and antimicrobial resistance of Vibrio species isolated from diseased marine fish in South China. Sci. Rep. 2020, 10, 14329. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.; Amal, M.N.A.; Saad, M.Z.; Yasin, I.S.M.; Zulkiply, N.A.; Mustafa, M.; Nasruddin, N.S. Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet. Res. 2019, 15, 176. [Google Scholar] [CrossRef] [PubMed]
- Xedzro, C.; Shimamoto, T.; Shimamoto, T. Antimicrobial resistance and genotypic attributes of virulence among Vibrio spp. isolated from Japanese retail seafood. J. Agric. Food Res. 2024, 18, 101449. [Google Scholar] [CrossRef]
- Kumar, A.; Das, B.; Kumar, N. Vibrio pathogenicity island-1: The master determinant of cholera pathogenesis. Front. Cell. Infect. Microbiol. 2020, 10, 561296. [Google Scholar] [CrossRef] [PubMed]
- Ellison, C.K.; Dalia, T.N.; Vidal Ceballos, A.; Wang, J.C.; Biais, N.; Brun, Y.V.; Dalia, A.B. Retraction of DNA-bound type IV competence pili initiates DNA uptake during natural transformation in Vibrio cholerae. Nat. Microbiol. 2018, 3, 773–780. [Google Scholar] [CrossRef]
- Hughes, H.Q.; Floyd, K.A.; Hossain, S.; Anantharaman, S.; Kysela, D.T.; Zöldi, M.; Barna, L.; Yu, Y.; Kappler, M.P.; Dalia, T.N.; et al. Nitric oxide stimulates type IV MSHA pilus retraction in Vibrio cholerae via activation of the phosphodiesterase CdpA. Proc. Natl. Acad. Sci. USA 2022, 119, e2108349119. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhong, Y.; Gu, X.; Yuan, J.; Saeed, A.F.; Wang, S. The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 2015, 6, 134247. [Google Scholar] [CrossRef]
- Wang, S.X.; Zhang, X.H.; Zhong, Y.B.; Sun, B.G.; Chen, J.X. Genes encoding the Vibrio harveyi haemolysin (VHH)/thermolabile haemolysin (TLH) are widespread in vibrios. Wei Sheng Wu Xue Bao = Acta Microbiol. Sin. 2007, 47, 874–881. [Google Scholar]
- Klein, S.L.; Gutierrez West, C.K.; Mejia, D.M.; Lovell, C.R. Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other Vibrionaceae species isolated from a pristine estuary. Appl. Environ. Microbiol. 2014, 80, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Mookerjee, S.; Batabyal, P.; Sarkar, M.H.; Palit, A. Seasonal prevalence of enteropathogenic Vibrio and their phages in the riverine estuarine ecosystem of South Bengal. PLoS ONE 2015, 10, e0137338. [Google Scholar] [CrossRef] [PubMed]
- Pettis, G.S.; Mukerji, A.S. Structure, function, and regulation of the essential virulence factor capsular polysaccharide of Vibrio vulnificus. Int. J. Mol. Sci. 2020, 21, 3259. [Google Scholar] [CrossRef]
- Gonciarz, R.L.; Renslo, A.R. Emerging role of ferrous iron in bacterial growth and host-pathogen interaction: New tools for chemical (micro)biology and antibacterial therapy. Curr. Opin. Chem. Biol. 2021, 61, 170–178. [Google Scholar] [CrossRef]
- Spiro, S.; Kilic, M.; Lewin, A.; Dobbin, S.; Thomson, A.J.; Moore, G.R. Interactions of heme and other metal ion complexes with the bacterial Fe-uptake regulatory protein and with bacterioferritin. In Iron Metabolism, Inorganic Biochemistry and Regulatory Mechanisms; Ferreira, G.C., Ed.; Wiley-VCH: Weinheim, Germany, 1999; pp. 211–226. [Google Scholar] [CrossRef]
- Eitinger, T.; Rodionov, D.A.; Grote, M.; Schneider, E. Canonical and ECF-type ATP-binding cassette importers in prokaryotes: Diversity in modular organization and cellular functions. FEMS Microbiol. Rev. 2011, 35, 3–67. [Google Scholar] [CrossRef]
- Abendroth, J.; Kreger, A.C.; Hol, W.G.J. The dimer formed by the periplasmic domain of EpsL from the Type 2 secretion system of Vibrio parahaemolyticus. J. Struct. Biol. 2009, 168, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Emch, M.; Yunus, M.; Sack, R.B. Are the environmental niches of Vibrio cholerae O139 different from those of Vibrio cholerae O1 El Tor? Int. J. Infect. Dis. 2001, 5, 214–219. [Google Scholar] [CrossRef]
- Bourgeois, J.; Lazinski, D.W.; Camilli, A. Identification of spacer and protospacer sequence requirements in the Vibrio cholerae Type I-E CRISPR/Cas system. mSphere 2020, 5, e00813-20. [Google Scholar] [CrossRef]
- Naser, I.B.; Hoque, M.M.; Nahid, M.A.; Tareq, T.M.; Rocky, M.K.; Faruque, S.M. Analysis of the CRISPR-Cas system in bacteriophages active on epidemic strains of Vibrio cholerae in Bangladesh. Sci. Rep. 2017, 7, 14880. [Google Scholar] [CrossRef]
- McDonald, N.D.; Regmi, A.; Morreale, D.P.; Borowski, J.D.; Boyd, E.F. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genom. 2019, 20, 105. [Google Scholar] [CrossRef]
- Zhang, E.; Zhou, W.; Zhou, J.; He, Z.; Zhou, Y.; Han, J.; Qu, D. CRISPR-Cas systems are present predominantly on chromosomes and their relationship with MEGs in Vibrio species. Arch. Microbiol. 2021, 204, 76. [Google Scholar] [CrossRef]
- Stern, A.; Keren, L.; Wurtzel, O.; Amitai, G.; Sorek, R. Self-targeting by CRISPR: Gene regulation or autoimmunity? Trends Genet. 2010, 26, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Letchumanan, V.; Chan, K.G.; Lee, L.H. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advanced molecular identification techniques. Front. Microbiol. 2014, 5, 705. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.; Chen, A.; Hasan, N.A.; Rashed, S.M.; Huq, A.; Colwell, R.R. Non-O1/non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay, Maryland. Appl. Environ. Microbiol. 2015, 81, 1909–1918. [Google Scholar] [CrossRef] [PubMed]

| Region | June | July | August | September | October | November | December | January | February | March | April | May |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R-1 | 28.0 | 22.3 | 34.8 | 21.1 | 20.0 | 16.0 | 8.7 | 11.5 | 9.2 | 10.1 | 11.4 | 15.8 |
| SP-1 | 24.1 | 31.6 | 31.1 | 20.2 | 22.0 | 20.2 | 13.1 | 13.3 | 17.4 | 15.2 | 11.1 | 18.0 |
| R-2 | 20.8 | 23.7 | 24.9 | 20.5 | 18.0 | 14.5 | 12.1 | 11.2 | 8.8 | 10.6 | 13.2 | 15.4 |
| SP-2 | 21.8 | 25.3 | 29.8 | 19.9 | 23.0 | 23.0 | 14.2 | 9.1 | 17.6 | 14.6 | 12.7 | 19.1 |
| R-3 | 21.0 | 23.6 | 25.4 | 21.3 | 17.6 | 15.5 | 13.7 | 10.8 | 9.3 | 9.2 | 11.2 | 13.9 |
| SP-3 | 23.0 | 23.1 | 31.7 | 20.8 | 21.5 | 24.4 | 9.0 | 8.7 | 15.7 | 11.6 | 11.0 | 13.2 |
| R-4 | 20.6 | 23.1 | 26.2 | 21.7 | 20.6 | 17.0 | 12.0 | 10.9 | 8.4 | 10.1 | 12.2 | 17.5 |
| SP-4 | 23.2 | 25.0 | 29.7 | 19.3 | 21.0 | 20.0 | 8.0 | 11.0 | 2.6 | 9.9 | 11.1 | 19.8 |
| Strain Name | Sample Type | Region/Sales Point | Sampling Month | MALDI-TOF MS Results | Genome-Based Phylogeny Results |
|---|---|---|---|---|---|
| 1-TCBS-A | RTE-SM | SP-2 | June 2022 | V. harveyi | V. jasicida |
| 1-TCBS-B | RTE-SM | SP-2 | June 2022 | NI | V. barjaei |
| 1-TCBS-C | RTE-SM | SP-2 | June 2022 | V. alginolyticus | V. alginolyticus |
| 1-TCBS-D | RTE-SM | SP-2 | June 2022 | V. alginolyticus | V. alginolyticus |
| 3-TSA-A | RTE-SM | SP-3 | June 2022 | V. rumoiensis | V. alginolyticus |
| 4-MA-B | FRM | R-3 | June 2022 | V. rumoiensis | V. rumoiensis |
| 4-TSA-C | FRM | R-3 | June 2022 | V. alginolyticus | V. alginolyticus |
| 5-MA-A1 | FRM | R-4 | June 2022 | V. alginolyticus | V. diabolicus |
| 6-MA-B | FRM | R-1 | June 2022 | V. furnissii | V. furnissii |
| 11-TSA-B2 | FRM | R-1 | July 2022 | V. alginolyticus | V. alginolyticus |
| 14-MA-B | FRM | R-2 | July 2022 | V. rumoiensis | V. rumoiensis |
| 15-MA-B | RTE-SM | SP-3 | August 2022 | V. alginolyticus | V. diabolicus |
| 15-TSA-B2 | RTE-SM | SP-3 | August 2022 | V. alginolyticus | V. alginolyticus |
| 34-PA-B | FRM | R-1 | October 2022 | V. harveyi | V. owensii |
| 34-TSA-A | FRM | R-1 | October 2022 | V. alginolyticus | V. alginolyticus |
| Strains | GeneBank ID | Genome Size (bp) | Genome Coverage | No. Contigs | GC Content (%) | Total Genes | Protein-Coding Genes (CDSs) | rRNAs (5S, 16S, 23S) | tRNAs | ncRNAs | Pseudogenes a |
|---|---|---|---|---|---|---|---|---|---|---|---|
| V. jasicida 1-TCBS-A | JBIHSE000000000.1 | 6,106,211 | 117 | 5 | 45.0 | 5499 | 5253 | 12, 12, 11 | 129 | 4 | 78 |
| V. barjaei 1-TCBS-B | JBIHSF000000000.1 | 5,739,451 | 32 | 12 | 44.2 | 5278 | 5084 | 9, 10, 8 | 108 | 4 | 55 |
| V. alginolyticus 1-TCBS-C | JBIHSG000000000.1 | 5,270,144 | 141 | 6 | 44.5 | 4818 | 4519 | 13, 12, 12 | 129 | 4 | 129 |
| V. alginolyticus 1-TCBS-D | JBIHSH000000000.1 | 5,112,826 | 157 | 5 | 44.7 | 4656 | 4312 | 13, 12, 12 | 129 | 4 | 174 |
| V. alginolyticus 3-TSA-A | JBIHSI000000000.1 | 5,168,198 | 156 | 5 | 44.5 | 4685 | 4414 | 10, 9, 9 | 125 | 4 | 114 |
| V. rumoiensis 4-MA-B | JBIHSJ000000000.1 | 3,932,657 | 88 | 4 | 41.9 | 3578 | 3378 | 9, 8, 8 | 93 | 4 | 78 |
| V. alginolyticus 4-TSA-C | JBIHSK000000000.1 | 5,175,393 | 52 | 8 | 44.6 | 4698 | 4442 | 10, 9, 8 | 118 | 4 | 107 |
| V. diabolicus 5-MA-A1 | JBIHSL000000000.1 | 5,300,992 | 155 | 4 | 44.7 | 4913 | 4632 | 9, 9, 9 | 122 | 5 | 127 |
| V. furnissii 6-MA-B | JBIHSM000000000.1 | 5,087,888 | 160 | 5 | 50.4 | 4762 | 4503 | 8, 9, 9 | 109 | 4 | 120 |
| V. alginolyticus 11-TSA-B2 | JBIHSQ000000000.1 | 5,142,241 | 155 | 2 | 44.6 | 4685 | 4405 | 13, 11, 12 | 126 | 4 | 114 |
| V. rumoiensis 14-MA-B | JBIHSN000000000.1 | 3,992,184 | 146 | 5 | 41.9 | 3665 | 3453 | 9, 8, 8 | 93 | 5 | 89 |
| V. diabolicus 15-MA-B | JBIHSR000000000.1 | 5,149,397 | 156 | 2 | 44.8 | 4713 | 4389 | 11, 12, 12 | 128 | 4 | 157 |
| V. alginolyticus 15-TSA-B2 | JBIHSS000000000.1 | 5,117,107 | 155 | 3 | 44.6 | 4637 | 4358 | 12, 11, 12 | 126 | 4 | 114 |
| V. owensii 34-PA-B | JBIHSO000000000.1 | 6,001,683 | 44 | 12 | 45.6 | 5418 | 5210 | 7, 10, 9 | 124 | 4 | 54 |
| V. alginolyticus 34-TSA-A | JBIHSP000000000.1 | 5,186,208 | 283 | 3 | 44.6 | 4720 | 4268 | 13, 12, 12 | 127 | 4 | 284 |
| Probability of Being a Human Pathogen | Matched Pathogenic Families | Prediction | |
|---|---|---|---|
| V. jasicida 1-TCBS-A | 0.715 | 24 | Human pathogen |
| V. barjaei 1-TCBS-B | 0.457 | 10 | Non-human pathogen |
| V. alginolyticus 1-TCBS-C | 0.838 | 48 | Human pathogen |
| V. alginolyticus 1-TCBS-D | 0.830 | 44 | Human pathogen |
| V. alginolyticus 3-TSA-A | 0.825 | 40 | Human pathogen |
| V. rumoiensis 4-MA-B | 0.417 | 4 | Non-human pathogen |
| V. alginolyticus 4-TSA-C | 0.849 | 44 | Human pathogen |
| V. diabolicus 5-MA-A1 | 0.861 | 49 | Human pathogen |
| V. furnissii 6-MA-B | 0.756 | 37 | Human pathogen |
| V. alginolyticus 11-TSA-B2 | 0.849 | 50 | Human pathogen |
| V. rumoiensis 14-MA-B | 0.535 | 8 | Human pathogen |
| V. diabolicus 15-MA-B | 0.864 | 49 | Human pathogen |
| V. alginolyticus 15-TSA-B2 | 0.839 | 44 | Human pathogen |
| V. owensii 34-PA-B | 0.706 | 28 | Human pathogen |
| V. alginolyticus 34-TSA-A | 0.828 | 42 | Human pathogen |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yibar, A.; Duman, M.; Ay, H.; Ajmi, N.; Tasci, G.; Gurler, F.; Guler, S.; Morick, D.; Saticioglu, I.B. Genomic Insight into Vibrio Isolates from Fresh Raw Mussels and Ready-to-Eat Stuffed Mussels. Pathogens 2025, 14, 52. https://doi.org/10.3390/pathogens14010052
Yibar A, Duman M, Ay H, Ajmi N, Tasci G, Gurler F, Guler S, Morick D, Saticioglu IB. Genomic Insight into Vibrio Isolates from Fresh Raw Mussels and Ready-to-Eat Stuffed Mussels. Pathogens. 2025; 14(1):52. https://doi.org/10.3390/pathogens14010052
Chicago/Turabian StyleYibar, Artun, Muhammed Duman, Hilal Ay, Nihed Ajmi, Gorkem Tasci, Fatma Gurler, Sabire Guler, Danny Morick, and Izzet Burcin Saticioglu. 2025. "Genomic Insight into Vibrio Isolates from Fresh Raw Mussels and Ready-to-Eat Stuffed Mussels" Pathogens 14, no. 1: 52. https://doi.org/10.3390/pathogens14010052
APA StyleYibar, A., Duman, M., Ay, H., Ajmi, N., Tasci, G., Gurler, F., Guler, S., Morick, D., & Saticioglu, I. B. (2025). Genomic Insight into Vibrio Isolates from Fresh Raw Mussels and Ready-to-Eat Stuffed Mussels. Pathogens, 14(1), 52. https://doi.org/10.3390/pathogens14010052






