Infectious Laryngotracheitis Virus and Avian Metapneumovirus: A Comprehensive Review
Abstract
:1. Introduction
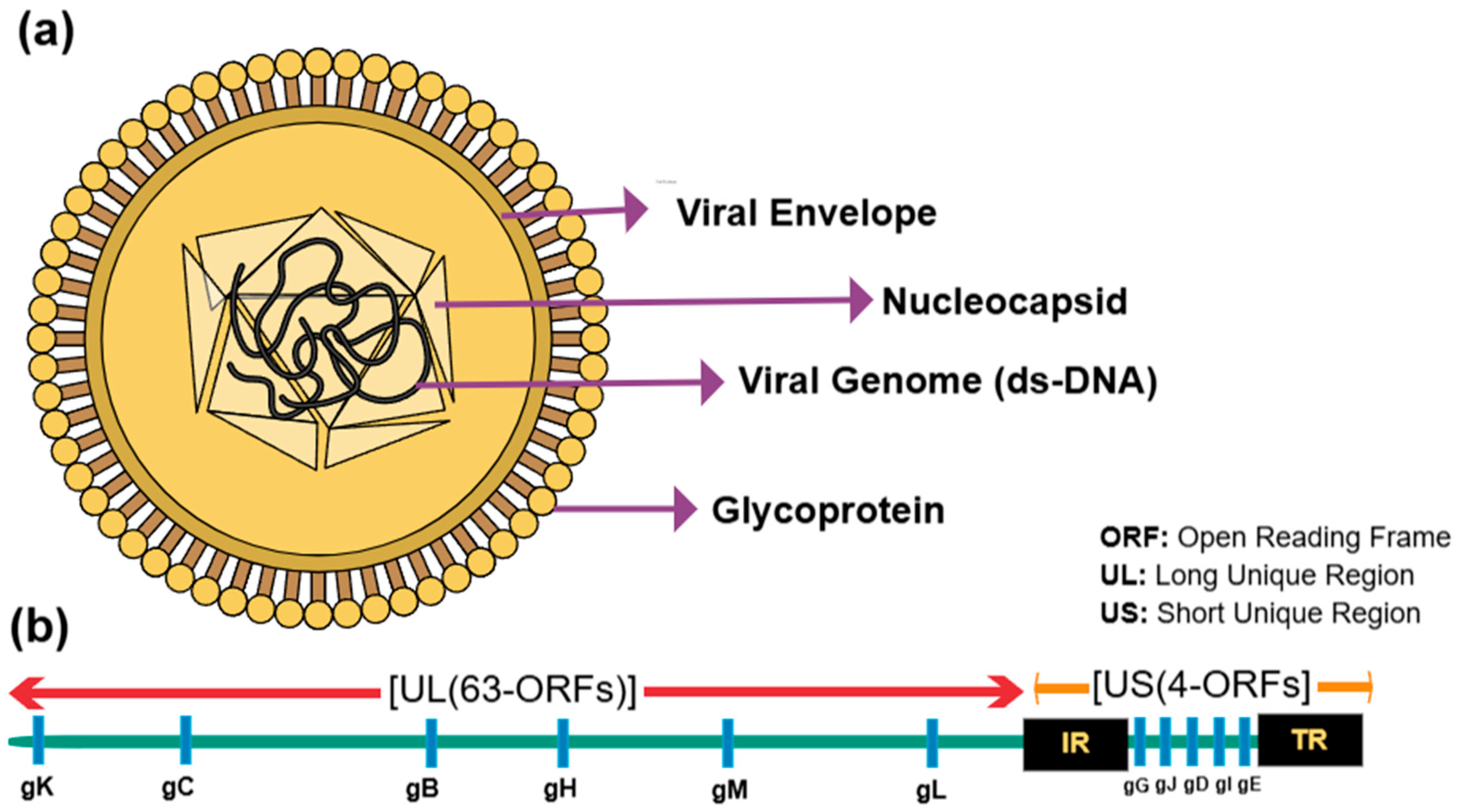
2. The Viral Structure and Classification of ILTV and aMPV
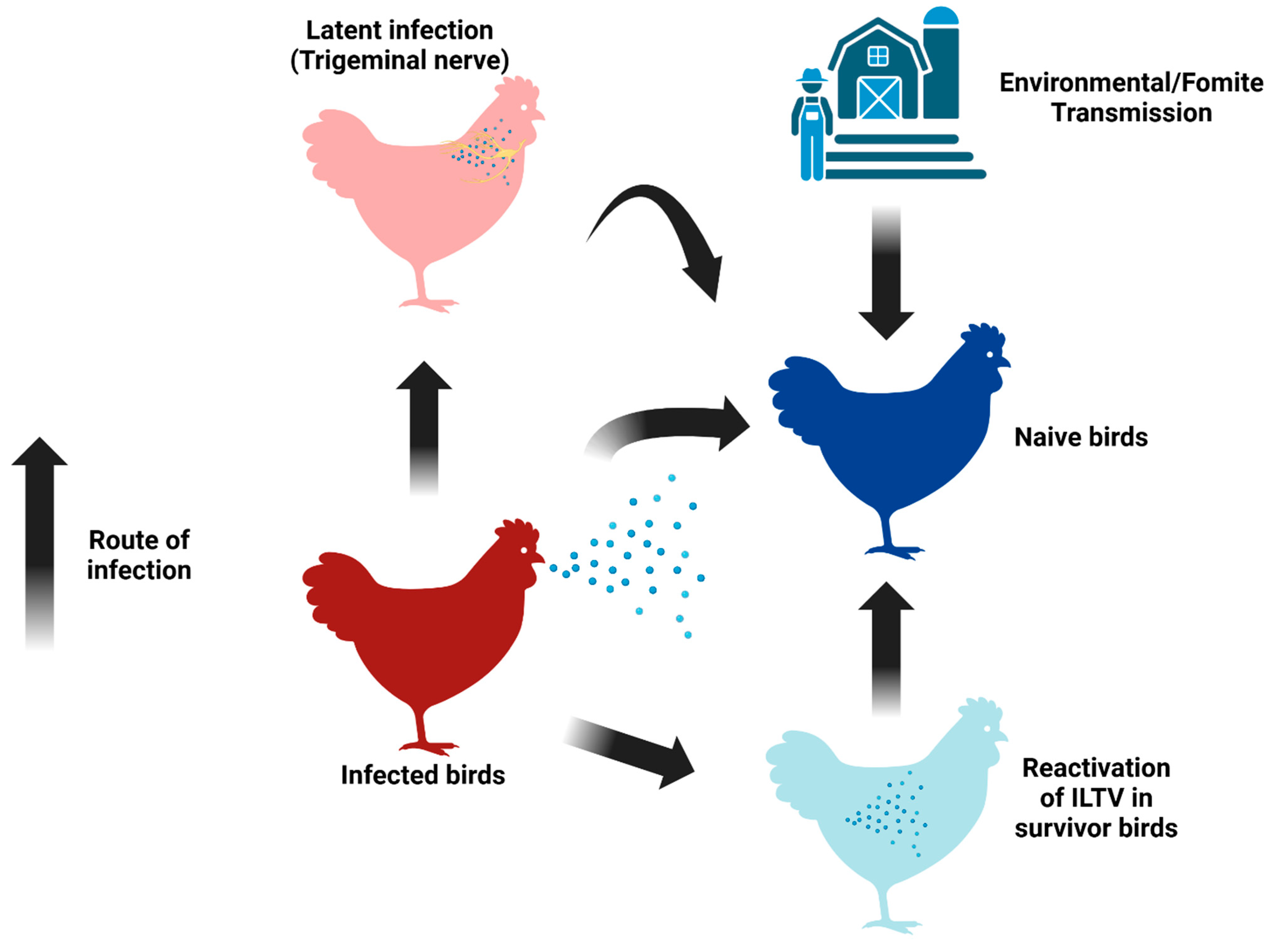
3. The Clinical Manifestations of ILTV and aMPV
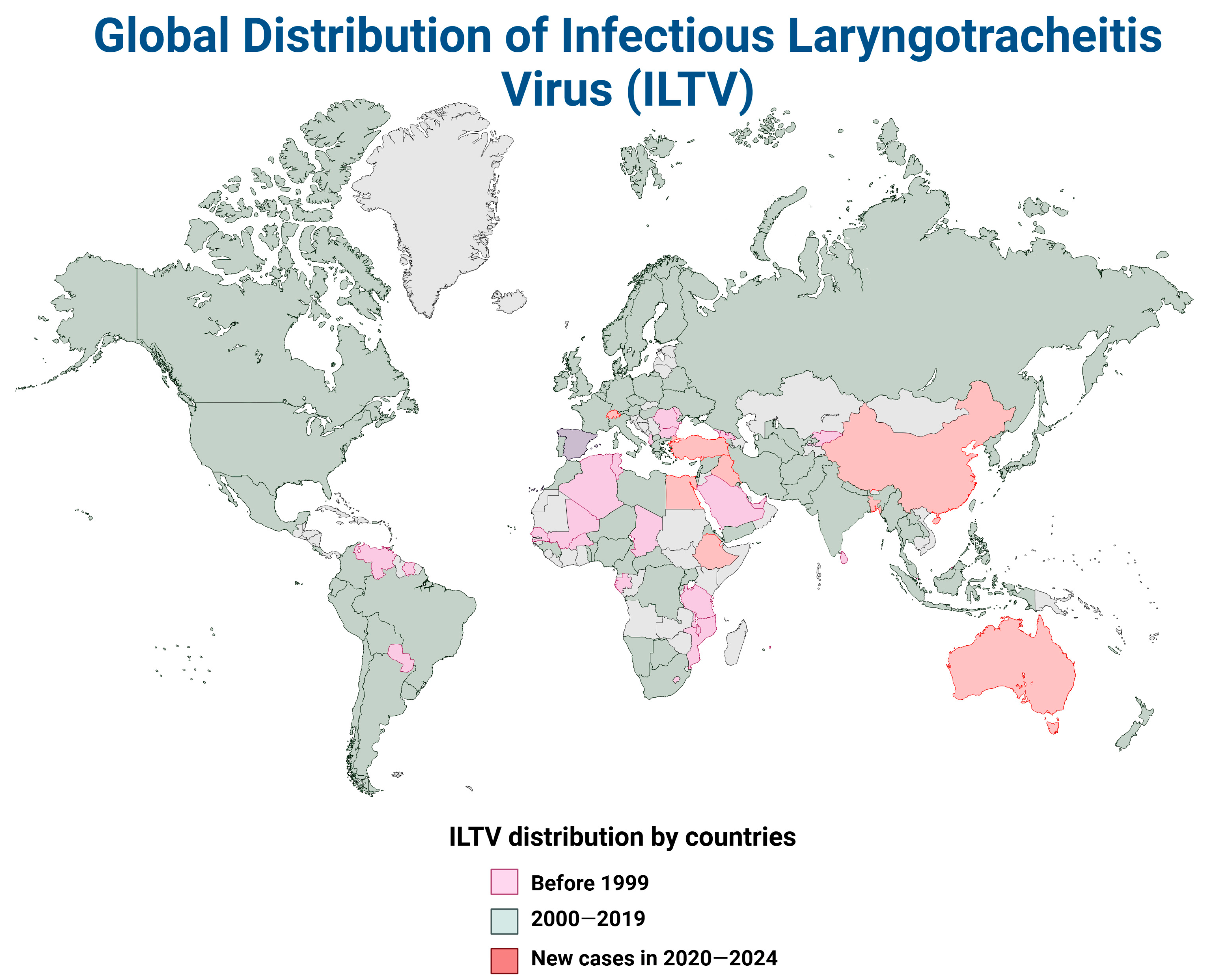
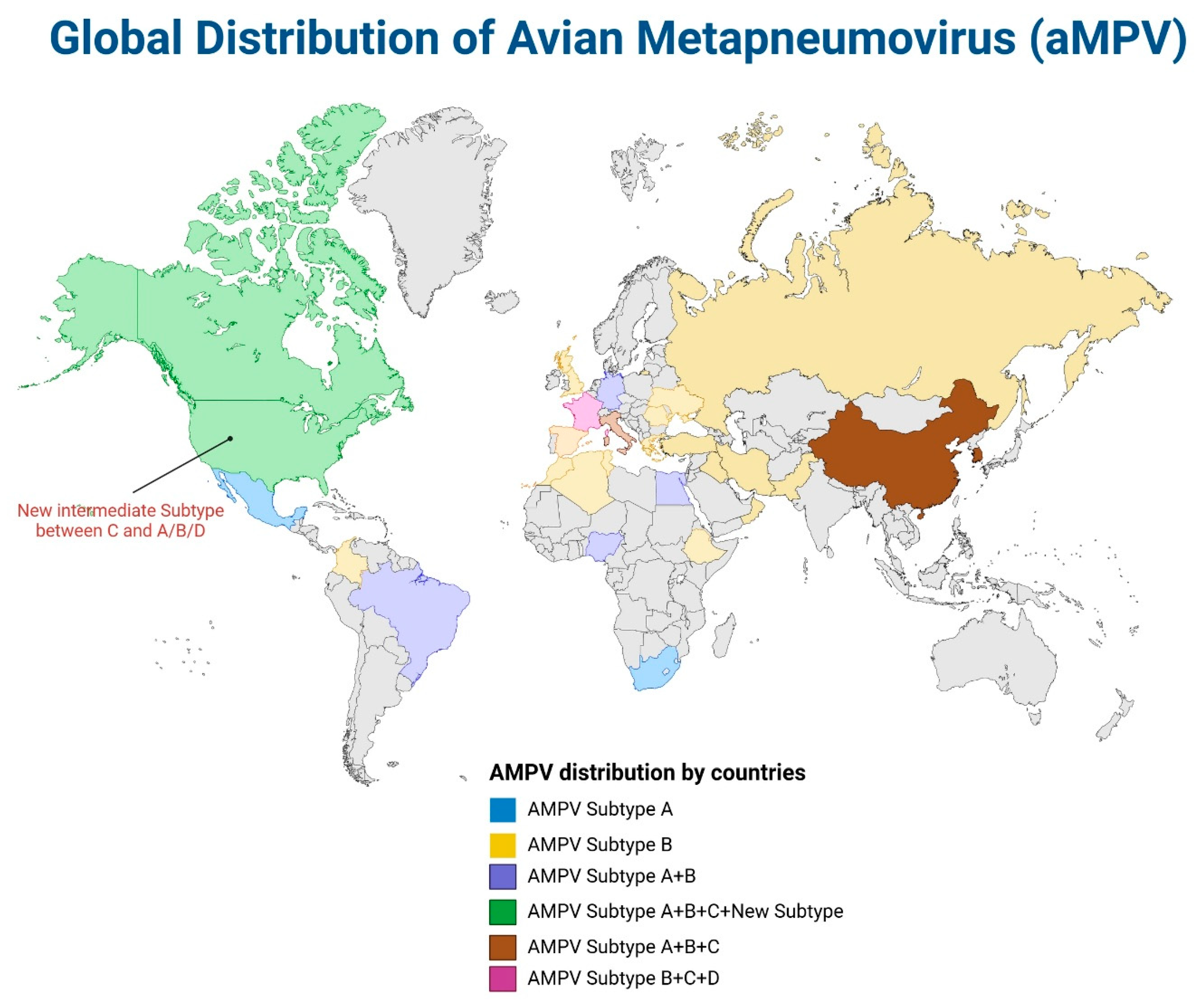
4. The Global Distribution of ILTV and aMPV
5. Vaccine Development for ILTV and aMPV
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jackwood, M.W. Review of infectious bronchitis virus around the world. Avian Dis. 2012, 56, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.J. An overview of the epidemiology of avian influenza. Vaccine 2007, 25, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.J.; Aldous, E.W.; Fuller, C.M. The long view: A selective review of 40 years of Newcastle disease research. Avian Pathol. 2012, 41, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Bagust, T.; Jones, R.; Guy, J. Avian infectious laryngotracheitis. Rev. Sci. Tech. (Int. Off. Epizoot.) 2000, 19, 483–492. [Google Scholar] [CrossRef]
- García, M. Current and future vaccines and vaccination strategies against infectious laryngotracheitis (ILT) respiratory disease of poultry. Vet. Microbiol. 2017, 206, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Menendez, K.R.; García, M.; Spatz, S.; Tablante, N.L. Molecular epidemiology of infectious laryngotracheitis: A review. Avian Pathol. 2014, 43, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Cover, M.S. The early history of infectious laryngotracheitis. Avian Dis. 1996, 40, 494–500. [Google Scholar] [CrossRef]
- Davison, A.J.; Eberle, R.; Ehlers, B.; Hayward, G.S.; McGeoch, D.J.; Minson, A.C.; Pellett, P.E.; Roizman, B.; Studdert, M.J.; Thiry, E. The order herpesvirales. Arch. Virol. 2009, 154, 171–177. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: An update. Virus Res. 2009, 143, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Gowthaman, V.; Kumar, S.; Koul, M.; Dave, U.; Murthy, T.G.K.; Munuswamy, P.; Tiwari, R.; Karthik, K.; Dhama, K.; Michalak, I.; et al. Infectious laryngotracheitis: Etiology, epidemiology, pathobiology, and advances in diagnosis and control—A comprehensive review. Vet. Q. 2020, 40, 140–161. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Veits, J.; Helferich, D.; Granzow, H.; Teifke, J.; Mettenleiter, T. Molecular biology of avian infectious laryngotracheitis virus. Vet. Res. 2007, 38, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Crawshaw, G.J.; Boycott, B.R. Infectious laryngotracheitis in peafowl and pheasants. Avian Dis. 1982, 26, 397–401. [Google Scholar] [CrossRef]
- Winterfield, R.; So, I.G. Susceptibility of turkeys to infectious laryngotracheitis. Avian Dis. 1968, 12, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Dufour-Zavala, L. Epizootiology of infectious laryngotracheitis and presentation of an industry control program. Avian Dis. 2008, 52, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bäyon-Auboyer, M.-H.; Arnauld, C.; Toquin, D.; Eterradossi, N. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J. Gen. Virol. 2000, 81, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Buys, S.; Preez, J.d. A preliminary report on the isolation of a virus causing sinusitis in turkeys in South Africa and attempts to attenuate the virus. Pluimvee Bull. 1989, 74. [Google Scholar]
- Perelman, B.; Meroz, M.; Samberg, Y. ‘Swollen head syndrome’ in broiler breeders in Israel. Vet. Rec. 1988, 123, 444. [Google Scholar] [CrossRef]
- Rojs, O.Z.; Stalcer, L. Swollen Head Syndrom (TRT): Current Situation in Broiler Breeders. In Proceedings of the VI Macedonian Poultry Days, Ohrid, North Macedonia, 6–9 May 1998. [Google Scholar]
- Hafez, H. Respiratory diseases in turkey: Serological surveillance for antibodies against Ornithobacterium rhinotracheale and turkey rhinotracheitis. In Proceedings of the 1th International Symposium on Turkey Disease, Berlin, Germany, 19–21 February 1998; pp. 138–145. [Google Scholar]
- Naylor, C.; Al-Ankari, A.; Al-Afaleq, A.; Bradbury, J.; Jones, R. Exacerbation of Mycoplasma gallisepticum infection in turkeys by rhinotracheitis virus. Avian Pathol. 1992, 21, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Marien, M.; Decostere, A.; Martel, A.; Chiers, K.; Froyman, R.; Nauwynck, H. Synergy between avian pneumovirus and Ornithobacterium rhinotracheale in turkeys. Avian Pathol. 2005, 34, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Cook, J.K.; Ellis, M.M.; Huggins, M.B. The pathogenesis of turkey rhinotracheitis virus in turkey poults inoculated with the virus alone or together with two strains of bacteria. Avian Pathol. 1991, 20, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Van de Zande, S.; Nauwynck, H.; De Jonghe, S.; Pensaert, M. Comparative pathogenesis of a subtype A with a subtype B avian pneumovirus in turkeys. Avian Pathol. 1999, 28, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Cook, J. Avian pneumovirus infections of turkeys and chickens. Vet. J. 2000, 160, 118–125. [Google Scholar] [CrossRef]
- Hassan, M.S.; Abdul-Careem, M.F. Avian viruses that impact table egg production. Animals 2020, 10, 1747. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Jirjis, F.; Kumar, M.; Njenga, M.; Shaw, D.; Noll, S.; Nagaraja, K.; Halvorson, D. Neonatal avian pneumovirus infection in commercial turkeys. Avian Dis. 2002, 46, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Williams, R.; Baxter-Jones, C.; Savage, C.; Wilding, G. Experimental infection of laying turkeys with rhinotracheitis virus: Distribution of virus in the tissues and serological response. Avian Pathol. 1988, 17, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Lwamba, H.C.; Bennett, R.S.; Lauer, D.C.; Halvorson, D.A.; Njenga, M.K. Characterization of avian metapneumoviruses isolated in the USA. Anim. Health Res. Rev. 2002, 3, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Watrach, A.; Hanson, L.; Watrach, M. The structure of infectious laryngotracheitis virus. Virology 1963, 21, 601–608. [Google Scholar] [CrossRef]
- Johnson, M.; Prideaux, C.; Kongsuwan, K.; Sheppard, M.; Fahey, K. Gallid herpesvirus 1 (infectious laryngotracheitis virus): Cloning and physical maps of the SA-2 strain. Arch. Virol. 1991, 119, 181–198. [Google Scholar] [CrossRef]
- Leib, D.; Bradbury, J.M.; Hart, C.; McCarthy, K. Genome isomerism in two alphaherpesviruses: Herpesvirus saimiri-1 (herpesvirus tamarinus) and avian infectious laryngotracheitis virus. Arch. Virol. 1987, 93, 287–294. [Google Scholar] [CrossRef]
- Fuchs, W.; Mettenleiter, T.C. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 1996, 77, 2221–2229. [Google Scholar] [CrossRef]
- Nadimpalli, M.; Lee, S.; Devlin, J.; Gilkerson, J.; Hartley, C. Impairment of infectious laryngotracheitis virus replication by deletion of the UL [-1] gene. Arch. Virol. 2017, 162, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Markham, P.F.; Markham, J.F.; Petermann, I.; Noormohammadi, A.H.; Browning, G.F.; Ficorilli, N.P.; Hartley, C.A.; Delvin, J.M. First complete genome sequence of infectious laryngotracheitis virus. BMC Genom. 2011, 12, 197. [Google Scholar] [CrossRef] [PubMed]
- Granzow, H.; Klupp, B.G.; Fuchs, W.; Veits, J.; Osterrieder, N.; Mettenleiter, T.C. Egress of alphaherpesviruses: Comparative ultrastructural study. J. Virol. 2001, 75, 3675–3684. [Google Scholar] [CrossRef]
- Hidalgo, H. Infectious laryngotracheitis: A review. Braz. J. Poult. Sci. 2003, 5, 157–168. [Google Scholar] [CrossRef]
- Thureen, D.R.; Keeler, C.L., Jr. Psittacid herpesvirus 1 and infectious laryngotracheitis virus: Comparative genome sequence analysis of two avian alphaherpesviruses. J. Virol. 2006, 80, 7863–7872. [Google Scholar] [CrossRef] [PubMed]
- Spear, P.G. Entry of alphaherpesviruses into cells. In Seminars in Virology; Elsevier: Amsterdam, The Netherlands, 1993; pp. 167–180. [Google Scholar]
- Bagust, T.J.; Johnson, M.A. Avian infectious laryngotracheitis: Virus-host interactions in relation to prospects for eradication. Avian Pathol. 1995, 24, 373–391. [Google Scholar] [CrossRef] [PubMed]
- York, J.J.; Sonza, S.; Brandon, M.; Fahey, K. Antigens of infectious laryngotracheitis herpesvirus defined by monoclonal antibodies. Arch. Virol. 1990, 115, 147–162. [Google Scholar] [CrossRef]
- Piccirillo, A.; Lavezzo, E.; Niero, G.; Moreno, A.; Massi, P.; Franchin, E.; Toppo, S.; Salata, C.; Palù, G. Full genome sequence-based comparative study of wild-type and vaccine strains of infectious laryngotracheitis virus from Italy. PLoS ONE 2016, 11, e0149529. [Google Scholar] [CrossRef]
- Pavlova, S.P.; Veits, J.; Blohm, U.; Maresch, C.; Mettenleiter, T.C.; Fuchs, W. In vitro and in vivo characterization of glycoprotein C-deleted infectious laryngotracheitis virus. J. Gen. Virol. 2010, 91, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Pringle, C. Virus taxonomy-San Diego 1998. Arch. Virol. 1998, 143, 1449. [Google Scholar] [CrossRef]
- Berthiaume, L.; Joncas, J.; Pavilanis, V. Comparative structure, morphogenesis and biological characteristics of the respiratory syncytial (RS) virus and the pneumonia virus of mice (PVM). Arch. Für Die Gesamte Virusforsch. 1974, 45, 39–51. [Google Scholar] [CrossRef]
- Compans, R.W.; Harter, D.H.; Choppin, P.W. Studies on Pneumonia Virus of Mice (Pvm) in Cell Culture: II. Structure and Morphogenesis of the Virus Particle. J. Exp. Med. 1967, 126, 267. [Google Scholar] [CrossRef]
- Collins, P.L.; Crowe, J.E., Jr.; Knipe, D.M.; Howley, P.M. Respiratory syncytial virus and metapneumovirus. In Virology, 5th ed.; LWW: London, UK, 2007; pp. 1601–1646. [Google Scholar]
- Easton, A.J.; Domachowske, J.B.; Rosenberg, H.F. Animal pneumoviruses: Molecular genetics and pathogenesis. Clin. Microbiol. Rev. 2004, 17, 390–412. [Google Scholar] [CrossRef] [PubMed]
- Feldman, S.A.; Audet, S.; Beeler, J.A. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 2000, 74, 6442–6447. [Google Scholar] [CrossRef] [PubMed]
- Hallak, L.K.; Collins, P.L.; Knudson, W.; Peeples, M.E. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 2000, 271, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Levine, S.; Klaiber-Franco, R.; Paradiso, P. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 1987, 68, 2521–2524. [Google Scholar] [CrossRef]
- Wertz, G.W.; Collins, P.L.; Huang, Y.; Gruber, C.; Levine, S.; Ball, L.A. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 1985, 82, 4075–4079. [Google Scholar] [CrossRef]
- Bastien, N.; Normand, S.; Taylor, T.; Ward, D.; Peret, T.C.; Boivin, G.; Anderson, L.J.; Li, Y. Sequence analysis of the N, P, M and F genes of Canadian human metapneumovirus strains. Virus Res. 2003, 93, 51–62. [Google Scholar] [CrossRef]
- van den Hoogen, B.G.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Analysis of the genomic sequence of a human metapneumovirus. Virology 2002, 295, 119–132. [Google Scholar] [CrossRef]
- Yu, Q.; Davis, P.; Barrett, T.; Binns, M.; Boursnell, M.; Cavanagh, D. Deduced amino acid sequence of the fusion glycoprotein of turkey rhinotracheitis virus has greater identity with that of human respiratory syncytial virus, a pneumovirus, than that of paramyxoviruses and morbilliviruses. J. Gen. Virol. 1991, 72, 75–81. [Google Scholar] [CrossRef]
- Zimmer, G.; Budz, L.; Herrler, G. Proteolytic activation of respiratory syncytial virus fusion protein: Cleavage at two furin consensus sequences. J. Biol. Chem. 2001, 276, 31642–31650. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, G.; Conzelmann, K.-K.; Herrler, G. Cleavage at the furin consensus sequence RAR/KR109 and presence of the intervening peptide of the respiratory syncytial virus fusion protein are dispensable for virus replication in cell culture. J. Virol. 2002, 76, 9218–9224. [Google Scholar] [CrossRef] [PubMed]
- Atreya, P.L.; Peeples, M.E.; Collins, P.L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J. Virol. 1998, 72, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Bossert, B.; Conzelmann, K.-K. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: A chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 2002, 76, 4287–4293. [Google Scholar] [CrossRef]
- Schlender, J.; Bossert, B.; Buchholz, U.; Conzelmann, K.-K. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 2000, 74, 8234–8242. [Google Scholar] [CrossRef]
- Dibben, O.; Thorpe, L.C.; Easton, A.J. Roles of the PVM M2-1, M2-2 and P gene ORF 2 (P-2) proteins in viral replication. Virus Res. 2008, 131, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, M.; Ferreira, H.L.; Munir, M.; Catelli, E. Avian metapneumovirus. In Mononegaviruses of Veterinary Importance Volume 2: Molecular Epidemiology and Control; CABI: Wallingford, UK, 2016; pp. 127–143. [Google Scholar]
- Biacchesi, S.; Skiadopoulos, M.H.; Boivin, G.; Hanson, C.T.; Murphy, B.R.; Collins, P.L.; Buchholz, U.J. Genetic diversity between human metapneumovirus subgroups. Virology 2003, 315, 1–9. [Google Scholar] [CrossRef]
- Brynes, A.; Williams, J.V. Small hydrophobic (SH) proteins of Pneumoviridae and Paramyxoviridae: Small but mighty. J. Virol. 2024, 98, e00809-24. [Google Scholar] [CrossRef]
- Biacchesi, S.; Skiadopoulos, M.H.; Yang, L.; Lamirande, E.W.; Tran, K.C.; Murphy, B.R.; Collins, P.L.; Buchholz, U.J. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: Deletion of G yields a promising vaccine candidate. J. Virol. 2004, 78, 12877–12887. [Google Scholar] [CrossRef]
- Biacchesi, S.; Pham, Q.N.; Skiadopoulos, M.H.; Murphy, B.R.; Collins, P.L.; Buchholz, U.J. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 2005, 79, 12608–12613. [Google Scholar] [CrossRef]
- Bendezu, J.; Morales Ruiz, S.; Montesinos, R.; Choque Guevara, R.; Rojas-Neyra, A.; Pauyac-Antezana, K.; Fernández-Díaz, M. Glycoprotein G (gG) production profile during infectious laryngotracheitis virus (ILTV) infection. PLoS ONE 2019, 14, e0219475. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, D.H.; Hazel, J.W.; Keeler, C.L., Jr. Identification and characterization of the infectious laryngotracheitis virus glycoprotein C gene. Virology 1994, 203, 336–343. [Google Scholar] [CrossRef]
- Kongsuwan, K.; Johnson, M.A.; Prideaux, C.T.; Sheppard, M. Identification of an infectious laryngotracheitis virus gene encoding an immunogenic protein with a predicted Mr of 32 kilodaltons. Virus Res. 1993, 29, 125–140. [Google Scholar] [CrossRef]
- Poulsen, D.J.; Keeler, C.L., Jr. Characterization of the assembly and processing of infectious laryngotracheitis virus glycoprotein B. J. Gen. Virol. 1997, 78, 2945–2951. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Browning, G.; Hartley, C.; Kirkpatrick, N.; Mahmoudian, A.; Noormohammadi, A.; Gilkerson, J. Glycoprotein G is a virulence factor in infectious laryngotracheitis virus. J. Gen. Virol. 2006, 87, 2839–2847. [Google Scholar] [CrossRef]
- Fuchs, W.; Wiesner, D.; Veits, J.; Teifke, J.P.; Mettenleiter, T.C. In vitro and in vivo relevance of infectious laryngotracheitis virus gJ proteins that are expressed from spliced and nonspliced mRNAs. J. Virol. 2005, 79, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Ponce-de-Leon, M.; Jiang, H.; Dubin, G.; Lubinski, J.M.; Eisenberg, R.J.; Cohen, G.H. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 1998, 72, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Backovic, M.; Klupp, B.G.; Rey, F.A.; Mettenleiter, T.C. Structure-based mutational analysis of the highly conserved domain IV of glycoprotein H of pseudorabies virus. J. Virol. 2012, 86, 8002–8013. [Google Scholar] [CrossRef]
- Di Giovine, P.; Settembre, E.C.; Bhargava, A.K.; Luftig, M.A.; Lou, H.; Cohen, G.H.; Eisenberg, R.J.; Krummenacher, C.; Carfi, A. Structure of herpes simplex virus glycoprotein D bound to the human receptor nectin-1. PLoS Pathog. 2011, 7, e1002277. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, A.; Nakanishi, H.; Kimura, K.; Matsubara, K.; Ozaki-Kuroda, K.; Katata, T.; Honda, T.; Kiyohara, Y.; Heo, K.; Higashi, M.; et al. Nectin: An adhesion molecule involved in formation of synapses. J. Cell Biol. 2002, 156, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, R.J.; Atanasiu, D.; Cairns, T.M.; Gallagher, J.R.; Krummenacher, C.; Cohen, G.H. Herpes virus fusion and entry: A story with many characters. Viruses 2012, 4, 800–832. [Google Scholar] [CrossRef] [PubMed]
- Zaichick, S.V.; Bohannon, K.P.; Smith, G.A. Alphaherpesviruses and the cytoskeleton in neuronal infections. Viruses 2011, 3, 941–981. [Google Scholar] [CrossRef] [PubMed]
- Shukla, N.D.; Tiwari, V.; Valyi-Nagy, T. Nectin-1-specific entry of herpes simplex virus 1 is sufficient for infection of the cornea and viral spread to the trigeminal ganglia. Mol. Vis. 2012, 18, 2711–2716. [Google Scholar]
- Kumar, V.; Yadav, K.; Kumar, R.; Chaudhary, N.; Kumar, S. Glycoprotein D peptide-based diagnostic approach for the detection of avian infectious laryngotracheitis antibodies. Avian Pathol. 2019, 48, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Loncoman, C.A.; Hartley, C.A.; Coppo, M.J.; Vaz, P.K.; Diaz-Méndez, A.; Browning, G.F.; García, M.; Spatz, S.; Devlin, J.M. Genetic diversity of infectious laryngotracheitis virus during in vivo coinfection parallels viral replication and arises from recombination hot spots within the genome. Appl. Environ. Microbiol. 2017, 83, e01532-17. [Google Scholar] [CrossRef]
- Perez Contreras, A.; van Der Meer, F.; Checkley, S.; Joseph, T.; King, R.; Ravi, M.; Peters, D.; Fonseca, K.; Gagnon, C.A.; Provost, C.; et al. Analysis of whole-genome sequences of infectious laryngotracheitis virus isolates from poultry flocks in Canada: Evidence of recombination. Viruses 2020, 12, 1302. [Google Scholar] [CrossRef]
- Loncoman, C.A.; Hartley, C.A.; Coppo, M.J.; Vaz, P.K.; Diaz-Méndez, A.; Browning, G.F.; Lee, S.; Devlin, J.M. Development and application of a TaqMan single nucleotide polymorphism genotyping assay to study infectious laryngotracheitis virus recombination in the natural host. PLoS ONE 2017, 12, e0174590. [Google Scholar] [CrossRef] [PubMed]
- Spatz, S.; Volkening, J.; Keeler, C.; Kutish, G.; Riblet, S.; Boettger, C.; Clark, K.F.; Zsak, L.; Afonso, C.L.; Mundt, E.S.; et al. Comparative full genome analysis of four infectious laryngotracheitis virus (Gallid herpesvirus-1) virulent isolates from the United States. Virus Genes 2012, 44, 273–285. [Google Scholar] [CrossRef]
- Beard, P.M.; Taus, N.S.; Baines, J.D. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 2002, 76, 4785–4791. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Weller, S.K. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J. Virol. 1992, 66, 469–479. [Google Scholar] [CrossRef]
- Smith, C.; Bates, P.; Rivera-Gonzalez, R.; Gu, B.; DeLuca, N. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J. Virol. 1993, 67, 4676–4687. [Google Scholar] [CrossRef]
- DeLuca, N.A.; Schaffer, P.A. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 1988, 62, 732–743. [Google Scholar] [CrossRef]
- Santander-Parra, S.H.; Nuñez, L.F.; Buim, M.R.; Ferreira, C.S.A.; Loncoman, C.A.; Ferreira, A.J.P. Detection and molecular characterization of infectious laryngotracheitis virus (ILTV) in chicken with respiratory signs in Brazil during 2015 and 2016. Braz. J. Microbiol. 2022, 53, 2223–2232. [Google Scholar] [CrossRef]
- Han, M.G.; Kim, S.J. Analysis of Korean strains of infectious laryngotracheitis virus by nucleotide sequences and restriction fragment length polymorphism. Vet. Microbiol. 2001, 83, 321–331. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Volkening, J.; Riblet, S.; Spatz, S. Genomic sequence analysis of the United States infectious laryngotracheitis vaccine strains chicken embryo origin (CEO) and tissue culture origin (TCO). Virology 2013, 440, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Veits, J.; Lüschow, D.; Kindermann, K.; Werner, O.; Teifke, J.P.; Mettenleiter, T.C.; Fuchs, W. Deletion of the non-essential UL0 gene of infectious laryngotracheitis (ILT) virus leads to attenuation in chickens, and UL0 mutants expressing influenza virus haemagglutinin (H7) protect against ILT and fowl plague. J. Gen. Virol. 2003, 84, 3343–3352. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, W.; Ziemann, K.; Teifke, J.P.; Werner, O.; Mettenleiter, T.C. The non-essential UL50 gene of avian infectious laryngotracheitis virus encodes a functional dUTPase which is not a virulence factor. J. Gen. Virol. 2000, 81, 627–638. [Google Scholar] [CrossRef]
- Collins, M.; Gough, R.; Alexander, D. Antigenic differentiation of avian pneumovirus isolates using polyclonal antisera and mouse monoclonal antibodies. Avian Pathol. 1993, 22, 469–479. [Google Scholar] [CrossRef]
- Juhasz, K.; Easton, A. Extensive sequence variation in the attachment (G) protein gene of avian pneumovirus: Evidence for two distinct subgroups. J. Gen. Virol. 1994, 75, 2873–2880. [Google Scholar] [CrossRef]
- Leib, D.; Bradbury, J.M.; Gaskell, R.M.; Hughes, C.S.; Jones, R. Restriction endonuclease patterns of some European and American isolates of avian infectious laryngotracheitis virus. Avian Dis. 1986, 30, 835–837. [Google Scholar] [CrossRef] [PubMed]
- Guy, J.S.; Barnes, H.J.; Munger, L.L.; Rose, L. Restriction endonuclease analysis of infectious laryngotracheitis viruses: Comparison of modified-live vaccine viruses and North Carolina field isolates. Avian Dis. 1989, 33, 316–323. [Google Scholar] [CrossRef]
- Oldoni, I.; García, M. Characterization of infectious laryngotracheitis virus isolates from the US by polymerase chain reaction and restriction fragment length polymorphism of multiple genome regions. Avian Pathol. 2007, 36, 167–176. [Google Scholar] [CrossRef] [PubMed]
- García, M.; Spatz, S.; Guy, J.S. Infectious laryngotracheitis. In Diseases of Poultry; Blackwell Publishing: Ames, IO, USA, 2013; pp. 161–179. [Google Scholar]
- Johnson, M.A.; Tyack, S.G.; Prideaux, C.; Kongsuwan, K.; Sheppard, M. Nucleotide sequence of infectious laryngotracheitis virus (gallid herpesvirus 1) ICP4 gene. Virus Res. 1995, 35, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Blakey, J.; Stoute, S.; Crossley, B.; Mete, A. Retrospective analysis of infectious laryngotracheitis in backyard chicken flocks in California, 2007–2017, and determination of strain origin by partial ICP4 sequencing. J. Vet. Diagn. Investig. 2019, 31, 350–358. [Google Scholar] [CrossRef]
- Priya, R.J.; Rao, G.V.S.; Pazhanivel, N.; Vijayarani, K.; Reetha, T.L.; Gowthaman, V.; Raja, P. Pathological diagnosis and genomic characterization of ICP4 gene of lnfectious laryngotracheitis virus (ILTV) isolates in clinically infected chicken in Tamil Nadu, India. Indian J. Anim. Res. 2023, 57, 770–776. [Google Scholar] [CrossRef]
- Zorman Rojs, O.; Dovč, A.; Krapež, U.; Žlabravec, Z.; Račnik, J.; Slavec, B. Detection of laryngotracheitis virus in poultry flocks with respiratory disorders in Slovenia. Viruses 2021, 13, 707. [Google Scholar] [CrossRef]
- Can-Sahna, K.; Abayli, H.; Ozbek, R.; Tonbak, S.; Bulut, H. Characterization of Infectious Laryngotracheitis Virus isolates from turkey by molecular and sequence analysis. Pak. Vet. J. 2020, 40, 337–342. [Google Scholar]
- Chacón, J.L.; Ferreira, A.J.P. Differentiation of field isolates and vaccine strains of infectious laryngotracheitis virus by DNA sequencing. Vaccine 2009, 27, 6731–6738. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, P.; Sukumar, K.; Raja, A.; Saravanan, S.; Srinivasan, P.; Thangavelu, A. Characterization of infectious laryngotracheitis virus isolates from laying hens during 2019–2020 outbreaks in Tamil Nadu, India. Arch. Virol. 2022, 167, 1819–1829. [Google Scholar] [CrossRef]
- Gough, A.; Pettit, J.; Gagnon, A.; Weber, L. An outbreak of infectious laryngotracheitis in commercial poultry flocks in Ontario. Can. J. Comp. Med. 1977, 41, 146–151. [Google Scholar] [PubMed]
- Härtle, S.; Vervelde, L.; Kaspers, B. The avian respiratory immune system. In Avian Immunology; Elsevier: Amsterdam, The Netherlands, 2022; pp. 327–341. [Google Scholar]
- Reddy, V.R.; Steukers, L.; Li, Y.; Fuchs, W.; Vanderplasschen, A.; Nauwynck, H.J. Replication characteristics of infectious laryngotracheitis virus in the respiratory and conjunctival mucosa. Avian Pathol. 2014, 43, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kong, C.; Cui, X.; Cui, H.; Shi, X.; Zhang, X.; Hu, S.; Hao, L.; Wang, Y. Detection of infectious laryngotracheitis virus by real-time PCR in naturally and experimentally infected chickens. PLoS ONE 2013, 8, e67598. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Egerton, J. Replication of infectious laryngotracheitis virus in chickens following vaccination. Aust. Vet. J. 1981, 57, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, N.C.; Mahmoudian, A.; Colson, C.A.; Devlin, J.M.; Noormohammadi, A.H. Relationship between mortality, clinical signs and tracheal pathology in infectious laryngotracheitis. Avian Pathol. 2006, 35, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Hitchner, S.; Fabricant, J.; Bagust, T. A fluorescent-antibody study of the pathogenesis of infectious laryngotracheitis. Avian Dis. 1977, 21, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Bennett, M.; Bradbury, J.; Gaskell, R.; Jones, R.; Jordan, F. Demonstration of sites of latency of infectious laryngotracheitis virus using the polymerase chain reaction. J. Gen. Virol. 1992, 73, 2415–2420. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.S.; LaRue, R.; Shaw, D.; Yu, Q.; Nagaraja, K.; Halvorson, D.A.; Njenga, M.K. A wild goose metapneumovirus containing a large attachment glycoprotein is avirulent but immunoprotective in domestic turkeys. J. Virol. 2005, 79, 14834–14842. [Google Scholar] [CrossRef] [PubMed]
- Velayudhan, B.T.; Yu, Q.; Estevez, C.N.; Nagaraja, K.V.; Halvorson, D.A. Glycoprotein gene truncation in avian metapneumovirus subtype C isolates from the United States. Virus Genes 2008, 37, 266–272. [Google Scholar] [CrossRef]
- Catelli, E.; De Marco, M.; Delogu, M.; Terregino, C.; Guberti, V. Serological evidence of avian pneumovirus infection in reared and free-living pheasants. Vet. Rec. 2001, 149, 56–58. [Google Scholar] [CrossRef] [PubMed]
- Cecchinato, M.; Lupini, C.; Silveira, F.; Listorti, V.; Mescolini, G.; Morandini, E.; Franzo, G.; Catelli, E. Molecular characterization of avian metapneumovirus from Guinea fowls (Numida meleagridis). Pak. Vet. J. 2018, 38, 419–423. [Google Scholar] [CrossRef]
- Jardine, C.; Parmley, E.; Buchanan, T.; Nituch, L.; Ojkic, D. Avian metapneumovirus subtype C in wild waterfowl in Ontario, Canada. Transbound. Emerg. Dis. 2018, 65, 1098–1102. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-J.; Nagaraja, K.V.; McComb, B.; Halvorson, D.A.; Jirjis, F.F.; Shaw, D.P.; Bruce, S.S.; Njenga, M.K. Isolation of avian pneumovirus from mallard ducks that is genetically similar to viruses isolated from neighboring commercial turkeys. Virus Res. 2002, 83, 207–212. [Google Scholar] [CrossRef]
- Turpin, E.; Stallknecht, D.; Slemons, R.; Zsak, L.; Swayne, D. Evidence of avian metapneumovirus subtype C infection of wild birds in Georgia, South Carolina, Arkansas and Ohio, USA. Avian Pathol. 2008, 37, 343–351. [Google Scholar] [CrossRef]
- Kaboudi, K.; Lachheb, J. Avian metapneumovirus infection in turkeys: A review on turkey rhinotracheitis. J. Appl. Poult. Res. 2021, 30, 100211. [Google Scholar] [CrossRef]
- Felippe, P.A.; Silva, L.H.A.d.; Santos, M.B.d.; Sakata, S.T.; Arns, C.W. Detection of and phylogenetic studies with avian metapneumovirus recovered from feral pigeons and wild birds in Brazil. Avian Pathol. 2011, 40, 445–452. [Google Scholar] [CrossRef]
- Graziosi, G.; Lupini, C.; Catelli, E. Disentangling the role of wild birds in avian metapneumovirus (aMPV) epidemiology: A systematic review and meta-analysis. Transbound. Emerg. Dis. 2022, 69, 3285–3299. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.A.; Allée, C.; Courtillon, C.; Szerman, N.; Lemaitre, E.; Toquin, D.; Mangart, J.M.; Amelot, M.; Eterradossi, N. Host specificity of avian metapneumoviruses. Avian Pathol. 2019, 48, 311–318. [Google Scholar] [CrossRef]
- Goyal, S.M.; Chiang, S.-J.; Dar, A.M.; Nagaraja, K.V.; Shaw, D.P.; Halvorson, D.A.; Kapur, V. Isolation of avian pneumovirus from an outbreak of respiratory illness in Minnesota turkeys. J. Vet. Diagn. Investig. 2000, 12, 166–168. [Google Scholar] [CrossRef] [PubMed]
- Jirjis, F.F.; Noll, S.; Halvorson, D.A.; Nagaraja, K.; Shaw, D. Pathogenesis of avian pneumovirus infection in turkeys. Vet. Pathol. 2002, 39, 300–310. [Google Scholar] [CrossRef]
- Pattison, M.; Chettle, N.; Randall, C.; Wyeth, P. Observations on swollen head syndrome in broiler and broiler breeder chickens. Vet. Rec. 1989, 125, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Mase, M.; Tanimura, N.; Yamaguchi, S.; Yuasa, N. Attempts to reproduce swollen head syndrome in specific pathogen-free chickens by inoculating with Escherichia coli and/or turkey rhinotracheitis virus. Avian Pathol. 1998, 27, 21–27. [Google Scholar] [CrossRef]
- Hafez, H. The role of pneumovirus in Swollen Head Syndrome of chicken. Arch. Für Geflügelkunde 1993, 57, 181–185. [Google Scholar]
- Shin, H.-J.; Cameron, K.T.; Jacobs, J.A.; Turpin, E.A.; Halvorson, D.A.; Goyal, S.M.; Nagaraja, K.V.; Kumar, M.C.; Lauer, D.C.; Seal, B.S. Molecular epidemiology of subgroup C avian pneumoviruses isolated in the United States and comparison with subgroup A and B viruses. J. Clin. Microbiol. 2002, 40, 1687–1693. [Google Scholar] [CrossRef]
- Kapczynski, D. Experimental infection of SPF laying turkeys with avian pneumovirus subtype C: Possible role of vertical transmission. In Proceedings of the American Association of Avian Pathologists, Minneasplis, MN, USA, 16–20 July 2005; p. 34. [Google Scholar]
- Escobar-Alfonso, S.; Alvarez-Mira, D.M.; Beltran-Leon, M.; Ramirez-Nieto, G.; Gomez, A.P. Avian Metapneumovirus Subtype B Circulation in Poultry and Wild Birds of Colombia. Pathogens 2024, 13, 882. [Google Scholar] [CrossRef]
- García, M.; Riblet, S.M. Characterization of infectious laryngotracheitis virus isolates: Demonstration of viral subpopulations within vaccine preparations. Avian Dis. 2001, 45, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Keeler, C.L., Jr.; Hazel, J.W.; Hastings, J.E.; Rosenberger, J.K. Restriction endonuclease analysis of Delmarva field isolates of infectious laryngotracheitis virus. Avian Dis. 1993, 37, 418–426. [Google Scholar] [CrossRef]
- Keller, L.H.; Benson, C.E.; Davison, S.; Eckroade, R.J. Differences among restriction endonuclease DNA fingerprints of Pennsylvania field isolates, vaccine strains, and challenge strains of infectious laryngotracheitis virus. Avian Dis. 1992, 36, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-C.; Lee, Y.-L.; Shien, J.-H.; Shieh, H.K. Rapid differentiation of vaccine strains and field isolates of infectious laryngotracheitis virus by restriction fragment length polymorphism of PCR products. J. Virol. Methods 1997, 66, 179–186. [Google Scholar] [CrossRef]
- Graham, D.; McLaren, I.; Calvert, V.; Torrens, D.; Meehan, B. RFLP analysis of recent Northern Ireland isolates of infectious laryngotracheitis virus: Comparison with vaccine virus and field isolates from England, Scotland and the Republic of Ireland. Avian Pathol. 2000, 29, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Hermann, S.; Stevens, M.J.; Sigrist, B.; Bilic, I.; Albini, S.; Wolfrum, N. Unveiling the genetic landscape of infectious laryngotracheitis virus in Switzerland: Evidence for vaccine-like and wild-type strains. Virology 2024, 600, 110217. [Google Scholar] [CrossRef]
- Alaraji, F.; Hammadi, H.; Abed, A.A.; Khudhair, Y.I. Molecular detection and phylogenetic tree of infectious laryngotracheitis virus in layers in Al-Diwaniyah province, Iraq. Vet. World 2019, 12, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Shooshtari, A.; Charkhkar, S. Differentiation of field isolates and vaccine strains of infectious laryngotracheitis virus by DNA sequencing. Afr. J. Microbiol. Res. 2011, 5, 4112–4117. [Google Scholar]
- Choi, E.-J.; La, T.-M.; Choi, I.-S.; Song, C.-S.; Park, S.-Y.; Lee, J.-B.; Lee, S.-W. Genotyping of infectious laryngotracheitis virus using allelic variations from multiple genomic regions. Avian Pathol. 2016, 45, 443–449. [Google Scholar] [CrossRef]
- Yan, Z.; Li, S.; Xie, Q.; Chen, F.; Bi, Y. Characterization of field strains of infectious laryngotracheitis virus in China by restriction fragment length polymorphism and sequence analysis. J. Vet. Diagn. Investig. 2016, 28, 46–49. [Google Scholar] [CrossRef]
- Neff, C.; Sudler, C.; Hoop, R. Characterization of western European field isolates and vaccine strains of avian infectious laryngotracheitis virus by restriction fragment length polymorphism and sequence analysis. Avian Dis. 2008, 52, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Magouz, A.; Medhat, S.; Abou Asa, S.; Desouky, A. Detection of infectious laryngotracheitis virus (Gallid herpesvirus-1) from clinically infected chickens in Egypt by different diagnostic methods. Iran. J. Vet. Res. 2018, 19, 194–201. [Google Scholar] [PubMed]
- Craig, M.I.; Rojas, M.F.; Van der Ploeg, C.A.; Olivera, V.; Vagnozzi, A.E.; Perez, A.M.; König, G. Molecular characterization and cluster analysis of field isolates of avian infectious laryngotracheitis virus from Argentina. Front. Vet. Sci. 2017, 4, 212. [Google Scholar] [CrossRef]
- Ojkic, D.; Swinton, J.; Vallieres, M.; Martin, E.; Shapiro, J.; Sanei, B.; Binnington, B. Characterization of field isolates of infectious laryngotracheitis virus from Ontario. Avian Pathol. 2006, 35, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Agnew-Crumpton, R.; Vaz, P.K.; Devlin, J.M.; O’Rourke, D.; Blacker-Smith, H.P.; Konsak-Ilievski, B.; Hartley, C.A.; Noormohammadi, A.H. Spread of the newly emerging infectious laryngotracheitis viruses in Australia. Infect. Genet. Evol. 2016, 43, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Nazir, S.; Yegoraw, A.A.; Charlesworth, R.P.; Williamson, S.; Sharpe, S.; Walkden-Brown, S.W.; Gerber, P.F. Marked differences in virulence of three Australian field isolates of infectious laryngotracheitis virus in meat and layer chickens. Avian Pathol. 2020, 49, 600–610. [Google Scholar] [CrossRef]
- Bayoumi, M.; El-Saied, M.; Amer, H.; Bastami, M.; Sakr, E.E.; El-Mahdy, M. Molecular characterization and genetic diversity of the infectious laryngotracheitis virus strains circulating in Egypt during the outbreaks of 2018 and 2019. Arch. Virol. 2020, 165, 661–670. [Google Scholar] [CrossRef]
- Kardoğan, Ö.; Sarıçam İnce, S. Molecular characterization and phylogenetic analysis of infectious laryngotracheitis virus isolates from commercial chicken flocks in Turkey. Arch. Virol. 2024, 169, 231. [Google Scholar] [CrossRef] [PubMed]
- Müştak, I.B.; Müştak, H.K. Circulation and Molecular Characterization of Infectious Laryngotracheitis Virus in Poultry Flocks with Respiratory Disorders in Turkey, 2018–2022. Avian Dis. 2024, 68, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Kamal, M.M.; Sadekuzzaman, M.; Parvin, K.; Haque, M.E.; Hayat, S.; Islam, M.A.; Khatun, M.M.; Siddique, M.P.; Nahar, S.S.; Khasruzzaman, A.K.M.; et al. Characterization of infectious laryngotracheitis virus isolated from commercial layer chickens in Bangladesh during the year 2021–2022. J. Adv. Vet. Anim. Res. 2024, 11, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Mulaw, A.; Demessie, Y.; Fagbohun, O.A.; Ayelet, G.; Shite, A.; Bitew, M. Seroprevalence, Risk Factors, and Molecular Detection of Infectious Laryngotracheitis in the Poultry of Western Amhara, Ethiopia. Acta Sci. Vet. Sci. 2024, 6, 15–22. [Google Scholar]
- Hong, X.; Zhang, H.; Zhang, X.; Zhang, X.P.; Zhang, T. A meta-analysis for prevalence of infectious laryngotracheitis in chickens in mainland China in 1981–2022. BMC Vet. Res. 2024, 20, 142. [Google Scholar] [CrossRef]
- Naylor, C.; Shaw, K.; Britton, P.; Cavanagh, D. Appearance of type B avian pneumovirus in Great Britain. Avian Pathol. 1997, 26, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Van de Zande, S.; Nauwynck, H.; Cavanagh, D.; Pensaert, M. Infections and reinfections with avian pneumovirus subtype A and B on Belgian turkey farms and relation to respiratory problems. J. Vet. Med. Ser. B 1998, 45, 621–626. [Google Scholar] [CrossRef]
- PeenState University. Avian Metapneumovirus. Available online: https://extension.psu.edu/avian-metapneumovirus (accessed on 9 July 2024).
- USDA-APHIS. Avian Metapneumovirus Update. In Agriculture USDA; USDA: Washington, DC, USA, 2024. Available online: https://www.aphis.usda.gov/news/program-update/avian-metapneumovirus-update (accessed on 9 January 2024).
- Wang, Y.S.; Li, M.; Tell, L.A.; Baynes, R.E.; Davis, J.L.; Vickroy, T.W.; Riviere, J.E.; Lin, Z. Physiological parameter values for physiologically based pharmacokinetic models in food-producing animals. Part II: Chicken and turkey. J. Vet. Pharmacol. Ther. 2021, 44, 423–455. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, S.; Sid, H.; Rautenschlein, S. Avian metapneumovirus infection of chicken and turkey tracheal organ cultures: Comparison of virus–host interactions. Avian Pathol. 2015, 44, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Goraichuk, I.V.; Torchetti, M.K.; Killian, M.L.; Kapczynski, D.R.; Sary, K.; Kulkarni, A.; Suarez, D.L. Introduction of Avian metapneumovirus subtype A to the United States: Molecular insights and implications. Front. Microbiol. 2024, 15, 1428248. [Google Scholar] [CrossRef] [PubMed]
- Kariithi, H.M.; Christy, N.; Decanini, E.L.; Lemiere, S.; Volkening, J.D.; Afonso, C.L.; Suarez, D.L. Detection and genome sequence analysis of avian metapneumovirus subtype A viruses circulating in commercial chicken flocks in Mexico. Vet. Sci. 2022, 9, 579. [Google Scholar] [CrossRef] [PubMed]
- Al-Hasan, B.A.; Alhatami, A.O.; Abdulwahab, H.M.; Bustani, G.S.; Hameed, M.A.; Jawad, A.H. First report of Avian metapneumovirus type B in Iraqi broiler flocks with swollen head syndrome. Vet. World 2022, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Kariithi, H.M.; Volkening, J.D.; Alves, V.V.; Reis-Cunha, J.L.; Arantes, L.C.R.V.; Fernando, F.S.; Filho, T.F.; Martins, N.R.D.S.; Lemiere, S.; Neto, O.C.D.F.; et al. Complete Genome Sequences of Avian Metapneumovirus Subtype B Vaccine Strains from Brazil. Microbiol. Resour. Announc. 2023, 12, e00235-23. [Google Scholar] [CrossRef] [PubMed]
- Lachheb, J.; Bouslama, Z.; Nsiri, J.; Badr, C.; Al Gallas, N.; Souissi, N.; Imed Khazri, I.; Larbi, I.; Kaboudi, K.; Ghram, A. Phylogenetic and phylodynamic analyses of subtype-B metapneumovirus from chickens in Tunisia. Poult. Sci. 2023, 102, 102253. [Google Scholar] [CrossRef]
- Wang, J.; Hou, L.; Wei, L.; Yan, X.; Zhu, S.; Quan, R.; Li, Z.; Wang, D.; Jiang, H.; Song, J.; et al. Characterization of avain metapneumovirus subgroup C isolated from chickens in Beijing, China. Poult. Sci. 2023, 102, 102250. [Google Scholar] [CrossRef]
- Mernizi, A.; Kadiri, O.; Rius, J.L.C.; Bouslikhane, M.; Ghram, A.; Mohamed, M.; Catelli, E.; Nassik, S. Detection of Avian Metapneumovirus Subtypes A and B in Moroccan Broiler Farms. Iran. J. Vet. Med. 2024, 18, 479–488. [Google Scholar] [CrossRef]
- Apinda, N.; Witoonsatian, K.; Sangkakam, K.; Muenthaisong, A.; Sthitmatee, N.; Tadee, P. Seroprevalence of avian metapneumovirus (aMPV) among pullet and layer hens in Northern Thailand. Trop. Anim. Health Prod. 2024, 56, 362. [Google Scholar] [CrossRef] [PubMed]
- Canuti, M.; Kroyer, A.N.; Ojkic, D.; Whitney, H.G.; Robertson, G.J.; Lang, A.S. Discovery and characterization of novel RNA viruses in aquatic North American wild birds. Viruses 2019, 11, 768. [Google Scholar] [CrossRef] [PubMed]
- Retallack, H.; Clubb, S.; DeRisi, J.L. Genome sequence of a divergent avian metapneumovirus from a monk parakeet (Myiopsitta monachus). Microbiol. Resour. Announc. 2019, 8, e00284-19. [Google Scholar] [CrossRef] [PubMed]
- Hilbink, F.; Oei, H.; Van Roozelaar, D. Virulence of five live vaccines against avian infectious laryngotracheitis and their immunogenicity and spread after eyedrop or spray application. Vet. Q. 1987, 9, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Avila, A.; Oldoni, I.; Riblet, S.; García, M. Evaluation of the protection elicited by direct and indirect exposure to live attenuated infectious laryngotracheitis virus vaccines against a recent challenge strain from the United States. Avian Pathol. 2008, 37, 287–292. [Google Scholar] [CrossRef]
- Samberg, Y.; Cuperstein, E.; Bendheim, U.; Aronovici, I. The development of a vaccine against avian infectious laryngotracheitis IV. Immunization of chickens with a modified laryngotracheitis vaccine in the drinking water. Avian Dis. 1971, 15, 413–417. [Google Scholar] [CrossRef]
- Gelenczei, E.; Marty, E. Studies on a tissue-culture-modified infectious laryngotracheitis virus. Avian Dis. 1964, 8, 105–122. [Google Scholar] [CrossRef]
- Cook, J.K.; Ellis, M.M.; Dolby, C.A.; Holmes, H.C.; Finney, P.M.; Huggins, M.B. A live attenuated turkey rhinotracheitis virus Vaccine 1. Stability of the attenuated strain. Avian Pathol. 1989, 18, 511–522. [Google Scholar] [CrossRef]
- Cook, J.K.; Holmes, H.; Finney, P.; Dolby, C.A.; Ellis, M.M.; Huggins, M. A live attenuated turkey rhinotracheitis virus Vaccine 2. The use of the attenuated strain as an experimental Vaccine. Avian Pathol. 1989, 18, 523–534. [Google Scholar] [CrossRef]
- Banet-Noach, C.; Simanov, L.; Laham-Karam, N.; Perk, S.; Bacharach, E. Longitudinal survey of avian metapneumoviruses in poultry in Israel: Infiltration of field strains into vaccinated flocks. Avian Dis. 2009, 53, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Catelli, E.; Lupini, C.; Cecchinato, M.; Ricchizzi, E.; Brown, P.; Naylor, C.J. Field avian metapneumovirus evolution avoiding vaccine induced immunity. Vaccine 2010, 28, 916–921. [Google Scholar] [CrossRef]
- Heffels-Redmann, U.; Sommer, D.; Kaleta, E. VI International Symposium on Avian Corona-and Pneumoviruses and Complicating Pathogens, Rauischholzhausen, Germany, 14–17 June 2009; VVB Laufersweiler: Giessen, Germany, 2009. [Google Scholar]
- Ling, R.; Sinkovic, S.; Toquin, D.; Guionie, O.; Eterradossi, N.; Easton, A.J. Deletion of the SH gene from avian metapneumovirus has a greater impact on virus production and immunogenicity in turkeys than deletion of the G gene or M2-2 open reading frame. J. Gen. Virol. 2008, 89, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Davison, S.; Gingerich, E.N.; Casavant, S.; Eckroade, R.J. Evaluation of the efficacy of a live fowlpox-vectored infectious laryngotracheitis/avian encephalomyelitis vaccine against ILT viral challenge. Avian Dis. 2006, 50, 50–54. [Google Scholar] [CrossRef]
- Bublot, M.; Pritchard, N.; Swayne, D.E.; Selleck, P.; Karaca, K.; Suarez, D.L.; Audonnet, J.C.; Mickle, T.R. Development and use of fowlpox vectored vaccines for avian influenza. Ann. N. Y. Acad. Sci. 2006, 1081, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Vagnozzi, A.; Zavala, G.; Riblet, S.M.; Mundt, A.; García, M. Protection induced by commercially available live-attenuated and recombinant viral vector vaccines against infectious laryngotracheitis virus in broiler chickens. Avian Pathol. 2012, 41, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Basavarajappa, M.K.; Kumar, S.; Khattar, S.K.; Gebreluul, G.T.; Paldurai, A.; Samal, S.K. A recombinant Newcastle disease virus (NDV) expressing infectious laryngotracheitis virus (ILTV) surface glycoprotein D protects against highly virulent ILTV and NDV challenges in chickens. Vaccine 2014, 32, 3555–3563. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Li, Y.; Dimitrov, K.; Afonso, C.L.; Spatz, S.; Zsak, L. Genetic stability of a Newcastle disease virus vectored infectious laryngotracheitis virus vaccine after serial passages in chicken embryos. Vaccine 2020, 38, 925–932. [Google Scholar] [CrossRef]
- Zeng, Z.; He, Y.; Wang, Z.; Yao, L.; Li, L.; Shang, Y.; Wang, H.; Zhang, R.; Shao, H.; Luo, Q.; et al. Characterization of a recombinant thermostable Newcastle disease virus (NDV) expressing glycoprotein gB of infectious laryngotracheitis virus (ILTV) protects chickens against ILTV challenge. Viruses 2023, 15, 500. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, M.S.; Siddique, F.; Hussain, I.; Ahmad, S.; Rafique, A. Thermostable vaccines for Newcastle disease: A review. Worlds Poult. Sci. J. 2014, 70, 829–838. [Google Scholar] [CrossRef]
- Avakian, A.; Wakenell, P.; Bryan, T.; Schaeffer, J.; Williams, C.; Whitfill, C. In ovo administration of Marek’s disease vaccine: Importance of vaccine deposition site in the fertile egg. In Proceedings of the 51st Western Poultry Disease Conference, Puerto Vallarta, Mexico, 1–4 May 2002. [Google Scholar]
- Williams, C.; Zedek, A. Comparative field evaluations of in ovo applied technology. Poult. Sci. 2010, 89, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Mundt, A.; Mundt, E.; Hogan, R.J.; García, M. Glycoprotein J of infectious laryngotracheitis virus is required for efficient egress of infectious virions from cells. Microbiol. Soc. 2011, 92, 2586–2589. [Google Scholar] [CrossRef]
- Veits, J.; Mettenleiter, T.C.; Fuchs, W. Five unique open reading frames of infectious laryngotracheitis virus are expressed during infection but are dispensable for virus replication in cell culture. J. Gen. Virol. 2003, 84, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.; Spatz, S.; Cheng, Y.; Riblet, S.; Volkening, J.; Schneiders, G. Attenuation and protection efficacy of ORF C gene-deleted recombinant of infectious laryngotracheitis virus. J. Gen. Virol. 2016, 97, 2352–2362. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.M.; Viejo-Borbolla, A.; Browning, G.F.; Noormohammadi, A.H.; Gilkerson, J.R.; Alcami, A.; Hartley, C.A. Evaluation of immunological responses to a glycoprotein G deficient candidate vaccine strain of infectious laryngotracheitis virus. Vaccine 2010, 28, 1325–1332. [Google Scholar] [CrossRef] [PubMed]
- Helferich, D.; Veits, J.; Teifke, J.P.; Mettenleiter, T.C.; Fuchs, W. The UL47 gene of avian infectious laryngotracheitis virus is not essential for in vitro replication but is relevant for virulence in chickens. J. Gen. Virol. 2007, 88, 732–742. [Google Scholar] [CrossRef]
- Han, M.; Kweon, C.; Mo, I.; Kim, S. Pathogenicity and vaccine efficacy of a thymidine kinase gene deleted infectious laryngotracheitis virus expressing the green fluorescent protein gene. Arch. Virol. 2002, 147, 1017–1031. [Google Scholar] [CrossRef]
- Ali, S.A.; Almofti, Y.A.; Abd-Elrahman, K.A. Immunoinformatics approach for multiepitopes vaccine prediction against glycoprotein B of avian infectious laryngotracheitis virus. Adv. Bioinform. 2019, 2019, 1270485. [Google Scholar] [CrossRef]
- Ibrahim, M.J.; Ali, S.A.; Abd-elrahman, K.A.; Almofti, Y.A. Vaccinomic approach for multi epitopes vaccine from glycoprotein D of virulent strains of avian infectious laryngotracheitis virus. J. Appl. Environ. Microbiol. 2020, 8, 8–24. [Google Scholar]
- Catelli, E.; Cecchinato, M.; Savage, C.E.; Jones, R.C.; Naylor, C.J. Demonstration of loss of attenuation and extended field persistence of a live avian metapneumovirus vaccine. Vaccine 2006, 24, 6476–6482. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Roth, J.P.; Hu, H.; Estevez, C.N.; Zhao, W.; Zsak, L. Protection by recombinant Newcastle disease viruses (NDV) expressing the glycoprotein (G) of avian metapneumovirus (aMPV) subtype A or B against challenge with virulent NDV and aMPV. World J. Vaccines 2013, 2013, 39829. [Google Scholar] [CrossRef]
- Hu, H.; Roth, J.P.; Zsak, L.; Yu, Q. Engineered Newcastle disease virus expressing the F and G proteins of AMPV-C confers protection against challenges in turkeys. Sci. Rep. 2017, 7, 4025. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Roth, J.P.; Estevez, C.N.; Zsak, L.; Liu, B.; Yu, Q. Generation and evaluation of a recombinant Newcastle disease virus expressing the glycoprotein (G) of avian metapneumovirus subgroup C as a bivalent vaccine in turkeys. Vaccine 2011, 29, 8624–8633. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Yu, M.; Wang, S.; Chen, Y.; Bao, Y.; Liu, P.; Feng, X.; He, T.; Guo, R.; Zhang, T.; et al. A novel live attenuated vaccine candidate protects chickens against subtype B avian metapneumovirus. J. Integr. Agric. 2024, 23, 1658–1670. [Google Scholar] [CrossRef]
- Naylor, C.J.; Brown, P.A.; Edworthy, N.; Ling, R.; Jones, R.C.; Savage, C.E.; Easton, A.J. Development of a reverse-genetics system for Avian pneumovirus demonstrates that the small hydrophobic (SH) and attachment (G) genes are not essential for virus viability. J. Gen. Virol. 2004, 85, 3219–3227. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, J.; Mo, J. Infectious Laryngotracheitis Virus and Avian Metapneumovirus: A Comprehensive Review. Pathogens 2025, 14, 55. https://doi.org/10.3390/pathogens14010055
Mo J, Mo J. Infectious Laryngotracheitis Virus and Avian Metapneumovirus: A Comprehensive Review. Pathogens. 2025; 14(1):55. https://doi.org/10.3390/pathogens14010055
Chicago/Turabian StyleMo, Jongsuk, and Jongseo Mo. 2025. "Infectious Laryngotracheitis Virus and Avian Metapneumovirus: A Comprehensive Review" Pathogens 14, no. 1: 55. https://doi.org/10.3390/pathogens14010055
APA StyleMo, J., & Mo, J. (2025). Infectious Laryngotracheitis Virus and Avian Metapneumovirus: A Comprehensive Review. Pathogens, 14(1), 55. https://doi.org/10.3390/pathogens14010055





