Genomic Diversity, Virulome, and Resistome of Streptococcus agalactiae in Northeastern Brazil: Are Multi-Host Adapted Strains Rising?
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Identification
2.2. Serotyping
2.3. Antimicrobial Susceptibility Tests
2.4. Statistical Analysis
2.5. Analysis of Clonality
2.6. Genome Sequencing
2.7. Genome Analysis
3. Results
3.1. S. agalactiae Clonal Relationships, Serotype Distributions and Antibiotic Susceptibilities
3.2. S. agalactiae Genome Analysis
3.3. S. agalactiae Genome Phylogeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raabe, V.N.; Shane, A.L. Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 2018, 7, GPP3-0007-2018. [Google Scholar] [CrossRef] [PubMed]
- Madhi, S.A.; Anderson, A.S.; Absalon, J.; Radley, D.; Simon, R.; Jongihlati, B.; Strehlau, R.; Van Niekerk, A.M.; Izu, A.; Naidoo, N.; et al. Potential for Maternally Administered Vaccine for Infant Group B Streptococcus. N. Engl. J. Med. 2023, 389, 215–227. [Google Scholar] [CrossRef]
- Furfaro, L.L.; Chang, B.J.; Payne, M.S. Perinatal Streptococcus agalactiae Epidemiology and Surveillance Targets. Clin. Microbiol. Rev. 2018, 31, e00049-18. [Google Scholar] [CrossRef] [PubMed]
- Zastempowska, E.; Twarużek, M.; Grajewski, J.; Lassa, H. Virulence Factor Genes and Cytotoxicity of Streptococcus agalactiae Isolated from Bovine Mastitis in Poland. Microbiol. Spectr. 2022, 10, e02224-21. [Google Scholar] [CrossRef]
- Coleman, M.; Armistead, B.; Orvis, A.; Quach, P.; Brokaw, A.; Gendrin, C.; Sharma, K.; Ogle, J.; Merillat, S.; Dacanay, M.; et al. Hyaluronidase Impairs Neutrophil Function and Promotes Group B Streptococcus Invasion and Preterm Labor in Nonhuman Primates. mBio 2021, 12, e03115-20. [Google Scholar] [CrossRef]
- Buscetta, M.; Firon, A.; Pietrocola, G.; Biondo, C.; Mancuso, G.; Midiri, A.; Romeo, L.; Galbo, R.; Venza, M.; Venza, I.; et al. PbsP, a Cell Wall-Anchored Protein That Binds Plasminogen to Promote Hematogenous Dissemination of Group B Streptococcus. Mol. Microbiol. 2016, 101, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Safadi, R.A.; Amor, S.; Hery-Arnaud, G.; Spellerberg, B.; Lanotte, P.; Mereghetti, L.; Gannier, F.; Quentin, R.; Rosenau, A. Enhanced Expression of Lmb Gene Encoding Laminin-Binding Protein in Streptococcus agalactiae Strains Harboring IS1548 in scpB-Lmb Intergenic Region. PLoS ONE 2010, 5, e10794. [Google Scholar] [CrossRef]
- Tazi, A.; Disson, O.; Bellais, S.; Bouaboud, A.; Dmytruk, N.; Dramsi, S.; Mistou, M.-Y.; Khun, H.; Mechler, C.; Tardieux, I.; et al. The Surface Protein HvgA Mediates Group B Streptococcus Hypervirulence and Meningeal Tropism in Neonates. J. Exp. Med. 2010, 207, 2313–2322. [Google Scholar] [CrossRef]
- Konto-Ghiorghi, Y.; Mairey, E.; Mallet, A.; Duménil, G.; Caliot, E.; Trieu-Cuot, P.; Dramsi, S. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 2009, 5, e1000422. [Google Scholar] [CrossRef]
- Metcalf, B.J.; Chochua, S.; Gertz, R.E.; Hawkins, P.A.; Ricaldi, J.; Li, Z.; Walker, H.; Tran, T.; Rivers, J.; Mathis, S.; et al. Short-Read Whole Genome Sequencing for Determination of Antimicrobial Resistance Mechanisms and Capsular Serotypes of Current Invasive Streptococcus agalactiae Recovered in the USA. Clin. Microbiol. Infect. 2017, 23, 574.e7–574.e14. [Google Scholar] [CrossRef]
- Costa, N.S.; Rio-Tinto, A.; Pinto, I.B.F.; Dos Santos Silva Alvim, D.C.; de Assis Rocha, A.; Oliveira, L.M.A.; Botelho, A.C.N.; Fracalanzza, S.E.L.; Teixeira, L.M.; Rezende-Filho, J.; et al. Changes in Group B Streptococcus Colonization among Pregnant Women before and after the Onset of the COVID-19 Pandemic in Brazil. Pathogens 2022, 11, 1104. [Google Scholar] [CrossRef]
- Lopes, E.; Fernandes, T.; Machado, M.P.; Carriço, J.A.; Melo-Cristino, J.; Ramirez, M.; Martins, E.R.; the Portuguese Group for the Study of Streptococcal Infections. Increasing Macrolide Resistance among Streptococcus agalactiae Causing Invasive Disease in Non-Pregnant Adults Was Driven by a Single Capsular-Transformed Lineage, Portugal, 2009 to 2015. Eurosurveillance 2018, 23, 1700473. [Google Scholar] [CrossRef]
- Crespo-Ortiz, M.d.P.; Castañeda-Ramirez, C.R.; Recalde-Bolaños, M.; Vélez-Londoño, J.D. Emerging Trends in Invasive and Noninvasive Isolates of Streptococcus agalactiae in a Latin American Hospital: A 17-Year Study. BMC Infect. Dis. 2014, 14, 428. [Google Scholar] [CrossRef] [PubMed]
- ACOG Practice Bulletin. Management of Herpes in Pregnancy. Number 8 October 1999: Clinical Management Guidelines for Obstetrician-Gynecologists. Int. J. Gynecol. Obstet. 2000, 68, 165–173. [Google Scholar] [CrossRef]
- Kerdsin, A.; Hatrongjit, R.; Hamada, S.; Akeda, Y.; Gottschalk, M. Development of a Multiplex PCR for Identification of β-Hemolytic Streptococci Relevant to Human Infections and Serotype Distribution of Invasive Streptococcus agalactiae in Thailand. Mol. Cell. Probes 2017, 36, 10–14. [Google Scholar] [CrossRef]
- Imperi, M.; Pataracchia, M.; Alfarone, G.; Baldassarri, L.; Orefici, G.; Creti, R. A Multiplex PCR Assay for the Direct Identification of the Capsular Type (Ia to IX) of Streptococcus agalactiae. J. Microbiol. Methods 2010, 80, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Sanger-Bentley-Group/GBS-Typer-Sanger-Nf 2024 v 1.0.12. Available online: https://github.com/sanger-bentley-group/GBS-Typer-sanger-nf (accessed on 8 December 2024).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; Clinical and Laboratory Standard Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Santos-Sanches, I.; Chambel, L.; Tenreiro, R. Pulsed-Field Gel Electrophoresis (PFGE): Application in Population Structure Studies of Bovine Mastitis-Causing Streptococci. In Veterinary Infection Biology: Molecular Diagnostics and High-Throughput Strategies; Cunha, M.V., Inácio, J., Eds.; Springer: New York, NY, USA, 2015; pp. 323–334. ISBN 978-1-4939-2004-4. [Google Scholar]
- Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update | Nucleic Acids Research | Oxford Academic. Available online: https://academic.oup.com/nar/article/52/W1/W83/7676834 (accessed on 8 December 2024).
- Wick, R.R.; Holt, K.E. Polypolish: Short-Read Polishing of Long-Read Bacterial Genome Assemblies. PLoS Comput. Biol. 2022, 18, e1009802. [Google Scholar] [CrossRef] [PubMed]
- Center for Genomic Epidemiology. Available online: https://www.genomicepidemiology.org/services/ (accessed on 8 December 2024).
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Soares, S.C.; Geyik, H.; Ramos, R.T.J.; de Sá, P.H.C.G.; Barbosa, E.G.V.; Baumbach, J.; Figueiredo, H.C.P.; Miyoshi, A.; Tauch, A.; Silva, A.; et al. GIPSy: Genomic Island Prediction Software. J. Biotechnol. 2016, 232, 2–11. [Google Scholar] [CrossRef]
- Lannes-Costa, P.S.; Baraúna, R.A.; Ramos, J.N.; Veras, J.F.C.; Conceição, M.V.R.; Vieira, V.V.; de Mattos-Guaraldi, A.L.; Ramos, R.T.J.; Doran, K.S.; Silva, A.; et al. Comparative Genomic Analysis and Identification of Pathogenicity Islands of Hypervirulent ST-17 Streptococcus agalactiae Brazilian Strain. Infect. Genet. Evol. 2020, 80, 104195. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Nagano, N.; Nagano, Y.; Kimura, K.; Tamai, K.; Yanagisawa, H.; Arakawa, Y. Genetic Heterogeneity in Pbp Genes among Clinically Isolated Group B Streptococci with Reduced Penicillin Susceptibility. Antimicrob. Agents Chemother. 2008, 52, 4258–4267. [Google Scholar] [CrossRef] [PubMed]
- Moroi, H.; Kimura, K.; Kotani, T.; Tsuda, H.; Banno, H.; Jin, W.; Wachino, J.-I.; Yamada, K.; Mitsui, T.; Yamashita, M.; et al. Isolation of Group B Streptococcus with Reduced β-Lactam Susceptibility from Pregnant Women. Emerg. Microbes Infect. 2019, 8, 2–7. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, K.; Du, X.; Lu, Y.; Liao, L.; He, Z.; He, W. Translational Attenuation Mechanism of ErmB Induction by Erythromycin Is Dependent on Two Leader Peptides. Front. Microbiol. 2021, 12, 690744. [Google Scholar] [CrossRef]
- Iannelli, F.; Santoro, F.; Santagati, M.; Docquier, J.-D.; Lazzeri, E.; Pastore, G.; Cassone, M.; Oggioni, M.R.; Rossolini, G.M.; Stefani, S.; et al. Type M Resistance to Macrolides Is Due to a Two-Gene Efflux Transport System of the ATP-Binding Cassette (ABC) Superfamily. Front. Microbiol. 2018, 9, 1670. [Google Scholar] [CrossRef]
- Salah, A.N.; Elleboudy, N.S.; El-Housseiny, G.S.; Yassien, M.A. Cloning and Sequencing of lsaE Efflux Pump Gene from MDR Enterococci and Its Role in Erythromycin Resistance. Infect. Genet. Evol. 2021, 94, 105010. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.A.; Law, C.S.; Metcalf, B.J.; Chochua, S.; Jackson, D.M.; Westblade, L.F.; Jerris, R.; Beall, B.W.; McGee, L. Cross-Resistance to Lincosamides, Streptogramins A and Pleuromutilins in Streptococcus agalactiae Isolates from the USA. J. Antimicrob. Chemother. 2017, 72, 1886–1892. [Google Scholar] [CrossRef]
- Teatero, S.; Athey, T.B.T.; Van Caeseele, P.; Horsman, G.; Alexander, D.C.; Melano, R.G.; Li, A.; Flores, A.R.; Shelburne, S.A.; McGeer, A.; et al. Emergence of Serotype IV Group B Streptococcus Adult Invasive Disease in Manitoba and Saskatchewan, Canada, Is Driven by Clonal Sequence Type 459 Strains. J. Clin. Microbiol. 2015, 53, 2919–2926. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Leekitcharoenphon, P.; Hansen, D.S.; Nielsen, H.L.; Ellermann-Eriksen, S.; Kemp, M.; Røder, B.L.; Frimodt-Møller, N.; Søndergaard, T.S.; et al. One Day in Denmark: Comparison of Phenotypic and Genotypic Antimicrobial Susceptibility Testing in Bacterial Isolates From Clinical Settings. Front. Microbiol. 2022, 13, 804627. [Google Scholar] [CrossRef]
- Botelho, A.C.N.; Ferreira, A.F.M.; Fracalanzza, S.E.L.; Teixeira, L.M.; Pinto, T.C.A. A Perspective on the Potential Zoonotic Role of Streptococcus agalactiae: Searching for a Missing Link in Alternative Transmission Routes. Front. Microbiol. 2018, 9, 608. [Google Scholar] [CrossRef]
- Khan, U.B.; Portal, E.A.R.; Sands, K.; Lo, S.; Chalker, V.J.; Jauneikaite, E.; Spiller, O.B. Genomic Analysis Reveals New Integrative Conjugal Elements and Transposons in GBS Conferring Antimicrobial Resistance. Antibiotics 2023, 12, 544. [Google Scholar] [CrossRef]
- Bianchi-Jassir, F.; Paul, P.; To, K.-N.; Carreras-Abad, C.; Seale, A.C.; Jauneikaite, E.; Madhi, S.A.; Russell, N.J.; Hall, J.; Madrid, L.; et al. Systematic Review of Group B Streptococcal Capsular Types, Sequence Types and Surface Proteins as Potential Vaccine Candidates. Vaccine 2020, 38, 6682–6694. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J. Group B Streptococcus: Virulence Factors and Pathogenic Mechanism. Microorganisms 2022, 10, 2483. [Google Scholar] [CrossRef] [PubMed]
- Banks, C.; Lindbom, B.J.; Kitson, G.; Darsley, M.; Fischer, P.B. Preclinical Development of a Novel Group B Streptococcus (GBS) Vaccine Candidate for Maternal Immunization Based upon the Alpha-like Protein Family of GBS Surface Proteins (Alp). Birth Defects Res. 2023, 115, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Sigaúque, B.; Kobayashi, M.; Vubil, D.; Nhacolo, A.; Chaúque, A.; Moaine, B.; Massora, S.; Mandomando, I.; Nhampossa, T.; Bassat, Q.; et al. Invasive Bacterial Disease Trends and Characterization of Group B Streptococcal Isolates among Young Infants in Southern Mozambique, 2001–2015. PLoS ONE 2018, 13, e0191193. [Google Scholar] [CrossRef] [PubMed]
- Springman, A.C.; Lacher, D.W.; Waymire, E.A.; Wengert, S.L.; Singh, P.; Zadoks, R.N.; Davies, H.D.; Manning, S.D. Pilus Distribution among Lineages of Group b Streptococcus: An Evolutionary and Clinical Perspective. BMC Microbiol. 2014, 14, 159. [Google Scholar] [CrossRef]
- Kallio, A.; Sepponen, K.; Hermand, P.; Denoël, P.; Godfroid, F.; Melin, M. Role of Pht Proteins in Attachment of Streptococcus Pneumoniae to Respiratory Epithelial Cells. Infect. Immun. 2014, 82, 1683–1691. [Google Scholar] [CrossRef]
- de Aguiar, E.L.; Mariano, D.C.B.; Viana, M.V.C.; Benevides, L.d.J.; de Souza Rocha, F.; de Castro Oliveira, L.; Pereira, F.L.; Dorella, F.A.; Leal, C.A.G.; de Carvalho, A.F.; et al. Complete Genome Sequence of Streptococcus agalactiae Strain GBS85147 Serotype of Type Ia Isolated from Human Oropharynx. Stand. Genom. Sci. 2016, 11, 39. [Google Scholar] [CrossRef]
- Sapugahawatte, D.N.; Li, C.; Dharmaratne, P.; Zhu, C.; Yeoh, Y.K.; Yang, J.; Lo, N.W.S.; Wong, K.T.; Ip, M. Prevalence and Characteristics of Streptococcus agalactiae from Freshwater Fish and Pork in Hong Kong Wet Markets. Antibiotics 2022, 11, 397. [Google Scholar] [CrossRef]
- Xuan, H.; Xia, L.; Schwarz, S.; Jia, H.; Yao, X.; Wang, S.; Li, R.; Wei, J.; Li, Z.; Shao, D.; et al. Various Mobile Genetic Elements Carrying optrA in Enterococcus Faecium and Enterococcus Faecalis Isolates from Swine within the Same Farm. J. Antimicrob. Chemother. 2023, 78, 504–511. [Google Scholar] [CrossRef]
- Crestani, C.; Forde, T.L.; Lycett, S.J.; Holmes, M.A.; Fasth, C.; Persson-Waller, K.; Zadoks, R.N. The Fall and Rise of Group B Streptococcus in Dairy Cattle: Reintroduction Due to Human-to-Cattle Host Jumps? Microb. Genom. 2021, 7, 000648. [Google Scholar] [CrossRef] [PubMed]
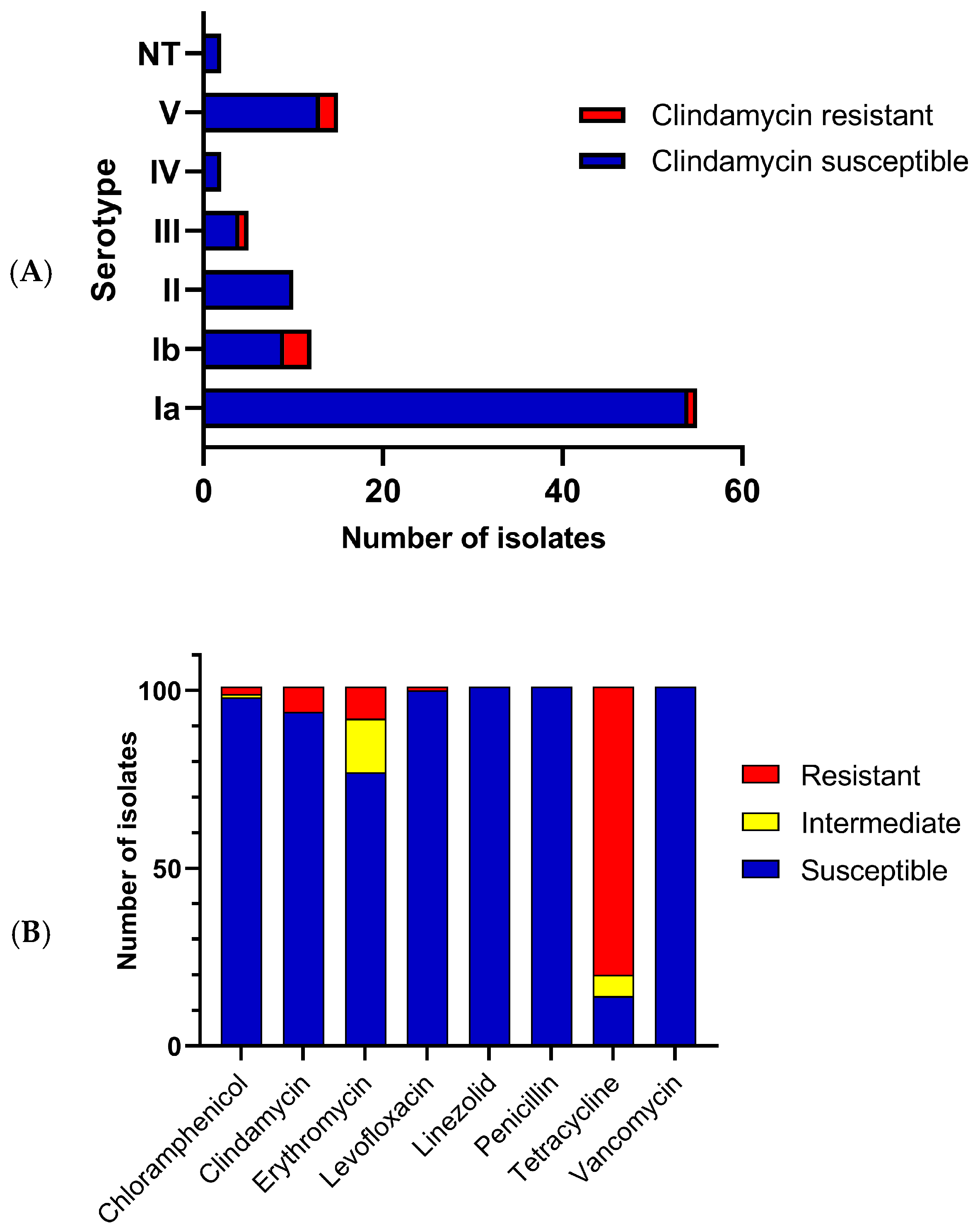
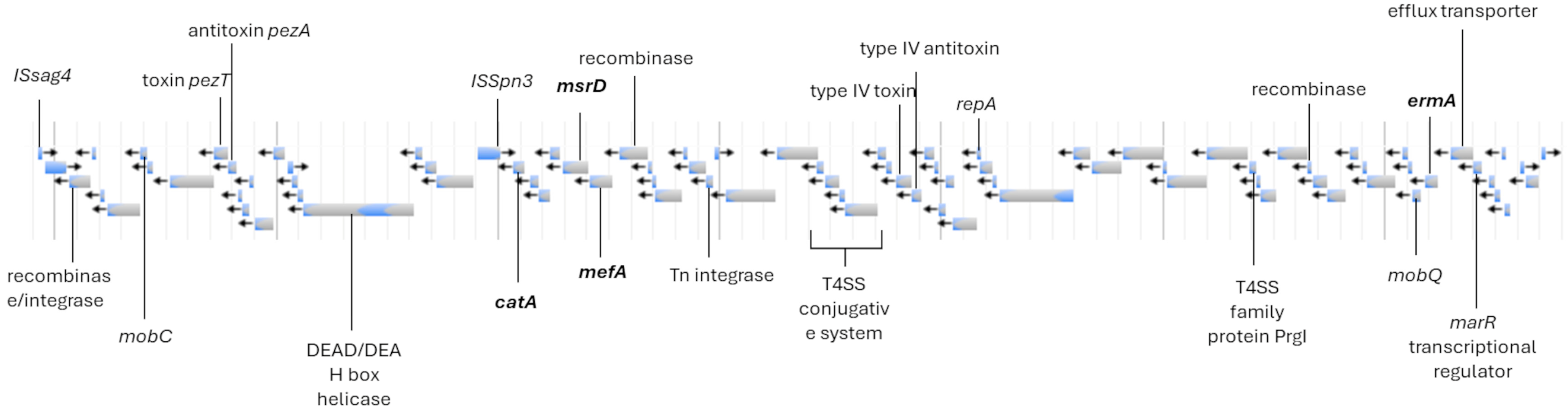
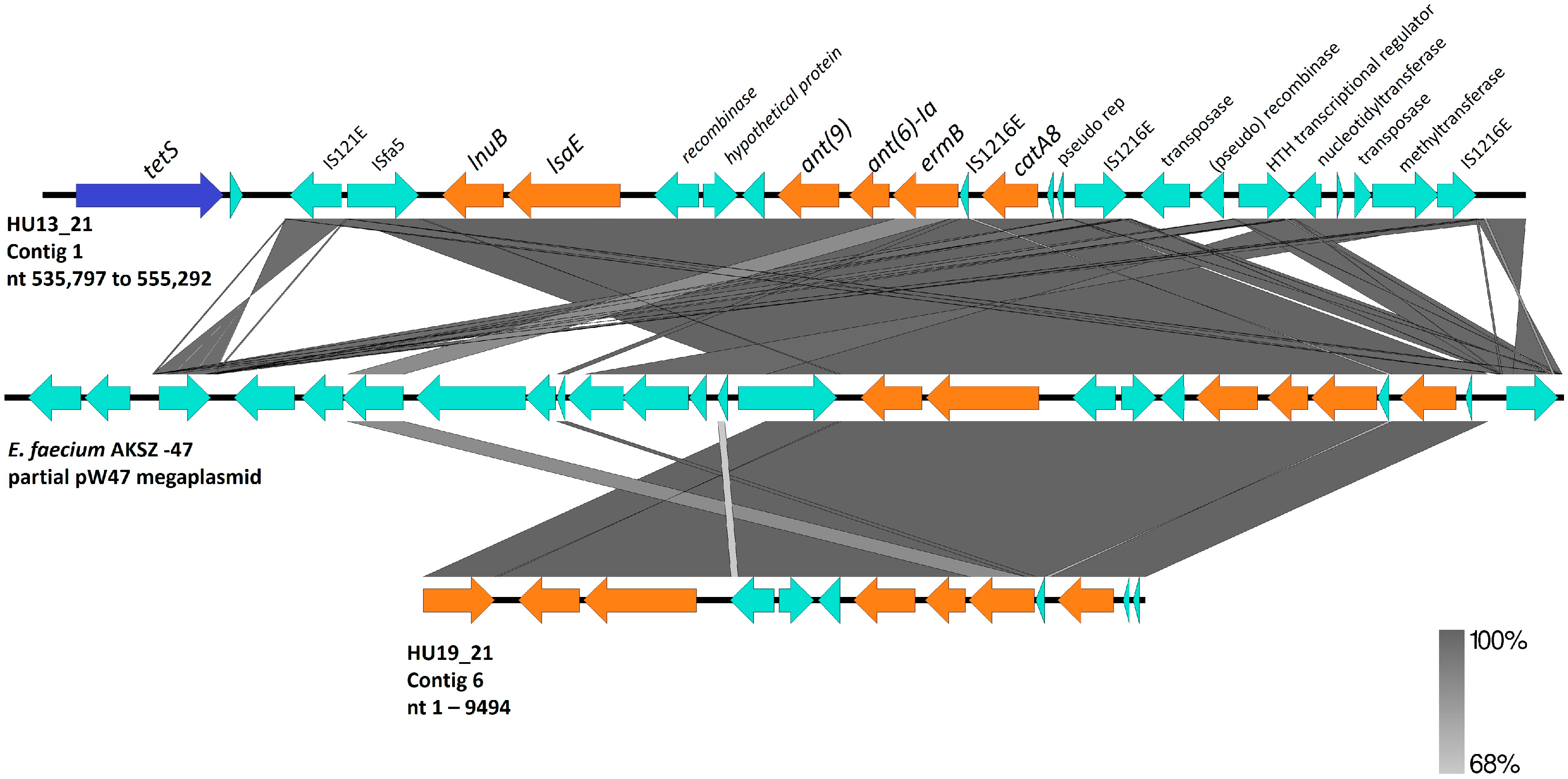

| Isolate | Source | ST | CC | Resistance Genes | Virulence Genes | Pilus Island | Access Number |
|---|---|---|---|---|---|---|---|
| HU05_19 | Urine | 28 | 19 | tetO | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip | PI-1 PI-2A2 | CP134689 |
| HU32_21 | Urine | 28 | 19 | tetM 1 | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-1 PI2A2 | CP135102 |
| MA06 | Urine | 19 | 19 | catA, ermA, gyrA (S81L), mefA, msrD parC (S79F), tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-2A2 | JASKHU000000000 |
| HU13_21 | Anovaginal | 196 | 459 | ant(6)-Ia, cat, ermB 2, lnuB 3, lsaE, tetS | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-1 | CP135101 |
| MA15 | Vaginal | 196 | 459 | ermA 2, mefA, msrD 3, tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-1 | CP135104 |
| HU19_21 | Anovaginal | 1983 | 103 | ant(6)-Ia, catA8, ermB, lnu, lsaE, tetS | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, rib, sip, srr1 | PI-2B | JASKHP000000000 |
| HU36_21 | Anovaginal | 1983 | 103 | tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, pbsP, rib, sip, srr1 | PI-2B | CP135103 |
| MA12 | Tracheal | 103 | 103 | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, pbsP, rib, scpB, sip, srr1 | PI-2B | CP143101 | |
| HU60_21 | Anovaginal | 12 | 12 | tetM | bac, bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-1 PI-2A2 | JAWIIQ000000000 |
| MA01 | Vaginal | 23 | 23 | tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-2A1 | JASKHT000000000 |
| MA07 | Vaginal | 144 | 23 | tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB sip, srr1 | PI-2A1 | CP134688 |
| HU29_21 | Urine | 144 | 23 | tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-2A1 | JAZGQP000000000 |
| HU30_21 | Urine | 144 | 23 | mefA, msrD, tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-2A1 | JAZGQQ000000000 |
| HU62_21 | Anovaginal | 144 | 23 | tetM | bca, cfb, cspA, cylE, fbsA, fbsB, fbsC, hylB, lmb, pbsP, rib, scpB, sip, srr1 | PI-2A1 | JASKHS000000000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perez, V.P.; Torini, L.R.; Manieri, F.Z.; de Queiroz, S.B.; de Brito Gomes, J.I.A.; Santos Filho, L.; Campana, E.H.; de Oliveira, C.J.B.; Sousa, E.S.S.; Camargo, I.L.B.C. Genomic Diversity, Virulome, and Resistome of Streptococcus agalactiae in Northeastern Brazil: Are Multi-Host Adapted Strains Rising? Pathogens 2025, 14, 292. https://doi.org/10.3390/pathogens14030292
Perez VP, Torini LR, Manieri FZ, de Queiroz SB, de Brito Gomes JIA, Santos Filho L, Campana EH, de Oliveira CJB, Sousa ESS, Camargo ILBC. Genomic Diversity, Virulome, and Resistome of Streptococcus agalactiae in Northeastern Brazil: Are Multi-Host Adapted Strains Rising? Pathogens. 2025; 14(3):292. https://doi.org/10.3390/pathogens14030292
Chicago/Turabian StylePerez, Vinicius Pietta, Luciana Roberta Torini, Fernanda Zani Manieri, Suellen Bernardo de Queiroz, Jorhanna Isabelle Araujo de Brito Gomes, Lauro Santos Filho, Eloiza Helena Campana, Celso Jose Bruno de Oliveira, Eduardo Sergio Soares Sousa, and Ilana Lopes Baratella Cunha Camargo. 2025. "Genomic Diversity, Virulome, and Resistome of Streptococcus agalactiae in Northeastern Brazil: Are Multi-Host Adapted Strains Rising?" Pathogens 14, no. 3: 292. https://doi.org/10.3390/pathogens14030292
APA StylePerez, V. P., Torini, L. R., Manieri, F. Z., de Queiroz, S. B., de Brito Gomes, J. I. A., Santos Filho, L., Campana, E. H., de Oliveira, C. J. B., Sousa, E. S. S., & Camargo, I. L. B. C. (2025). Genomic Diversity, Virulome, and Resistome of Streptococcus agalactiae in Northeastern Brazil: Are Multi-Host Adapted Strains Rising? Pathogens, 14(3), 292. https://doi.org/10.3390/pathogens14030292







