Does the Hfq Protein Contribute to RNA Cargo Translocation into Bacterial Outer Membrane Vesicles?
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of Hfq-CTR
2.3. DsrA sRNA Sequence and Preparation
2.4. Preparation of Small Unilamellar Vesicles (SUVs)
2.5. AFM Imaging in Solution
3. Results
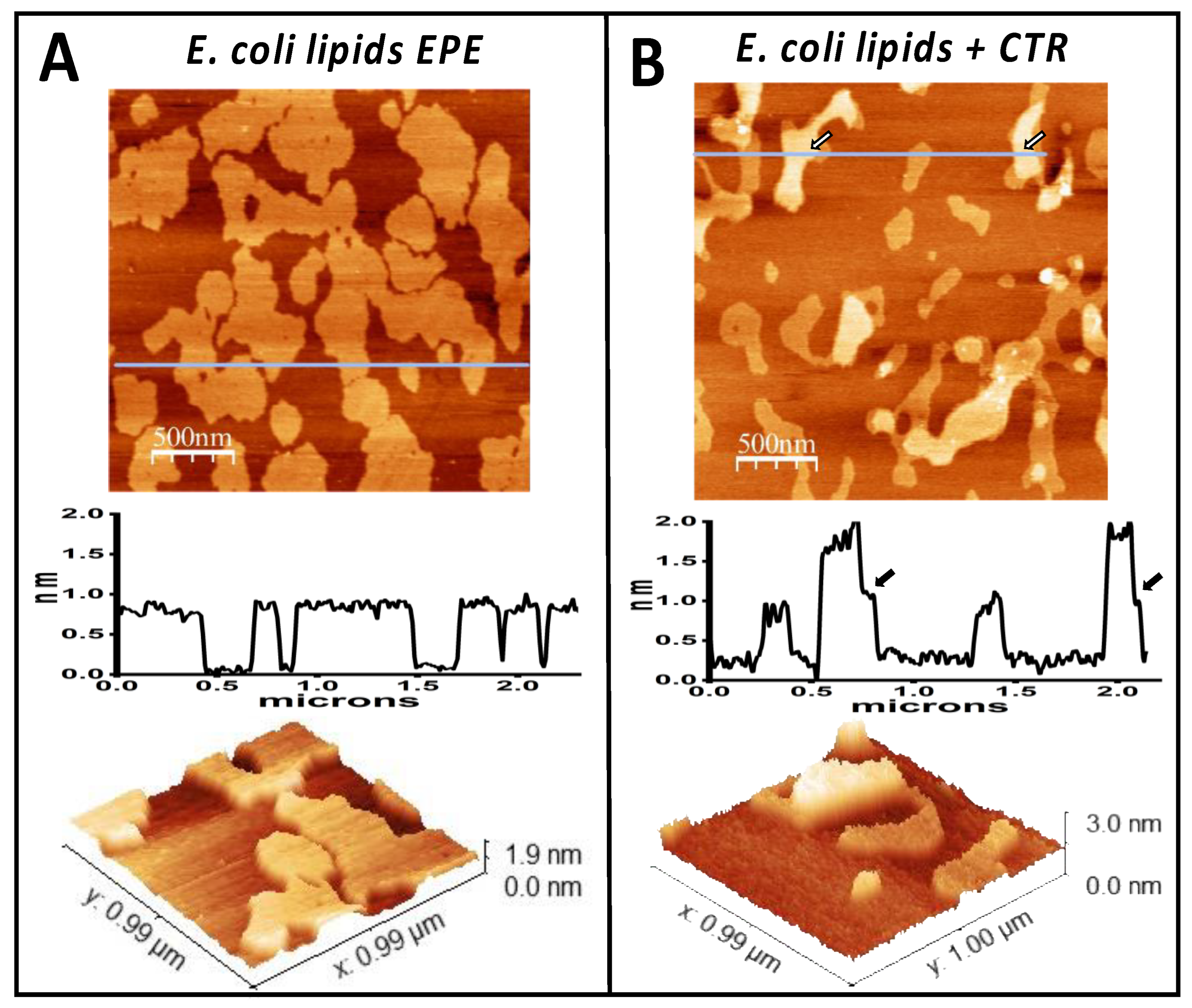
3.1. The Amyloid CTR Region of Hfq Binds to Membrane Microdomains
3.2. The Amyloid CTR Region of Hfq Triggers RNA Insertion in the Memrbane
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Papenfort, K.; Melamed, S. Small RNAs, Large Networks: Posttranscriptional Regulons in Gram-Negative Bacteria. Annu. Rev. Microbiol. 2023, 77, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Kavita, K.; de Mets, F.; Gottesman, S. New aspects of RNA-based regulation by Hfq and its partner sRNAs. Curr. Opin. Microbiol. 2018, 42, 53–61. [Google Scholar] [CrossRef]
- Gottesman, S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria. J. Biol. Chem. 2019, 294, 11685–11700. [Google Scholar] [CrossRef] [PubMed]
- Gelsinger, D.R.; DiRuggiero, J. The Non-Coding Regulatory RNA Revolution in Archaea. Genes 2018, 9, 141. [Google Scholar] [CrossRef]
- Svensson, S.L.; Sharma, C.M. Small RNAs in Bacterial Virulence and Communication. Microbiol. Spectr. 2016, 4, 169–212. [Google Scholar] [CrossRef]
- Pulvermacher, S.C.; Stauffer, L.T.; Stauffer, G.V. Role of the Escherichia coli Hfq protein in GcvB regulation of oppA and dppA mRNAs. Microbiology 2009, 155, 115–123. [Google Scholar] [CrossRef]
- Rice, J.B.; Vanderpool, C.K. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 2011, 39, 3806–3819. [Google Scholar] [CrossRef] [PubMed]
- Salvail, H.; Masse, E. Regulating iron storage and metabolism with RNA: An overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip. Rev. RNA 2012, 3, 26–36. [Google Scholar] [CrossRef]
- Majdalani, N.; Cunning, C.; Sledjeski, D.; Elliott, T.; Gottesman, S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 1998, 95, 12462–12467. [Google Scholar] [CrossRef]
- Melamed, S.; Adams, P.P.; Zhang, A.; Zhang, H.; Storz, G. RNA-RNA Interactomes of ProQ and Hfq Reveal Overlapping and Competing Roles. Mol. Cell 2020, 77, 411–425 e417. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Pearson, R.F.; Moller, T.; Valentin-Hansen, P.; Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: A bacterial Sm-like protein. Embo J. 2002, 21, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Link, T.M.; Valentin-Hansen, P.; Brennan, R.G. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA 2009, 106, 19292–19297. [Google Scholar] [CrossRef]
- Sauer, E.; Schmidt, S.; Weichenrieder, O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 9396–9401. [Google Scholar] [CrossRef]
- Dimastrogiovanni, D.; Frohlich, K.S.; Bandyra, K.J.; Bruce, H.A.; Hohensee, S.; Vogel, J.; Luisi, B.F. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. Elife 2014, 3, e05375. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.; Arluison, V.; Hohng, S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011, 39, 5131–5139. [Google Scholar] [CrossRef] [PubMed]
- De Lay, N.; Schu, D.J.; Gottesman, S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 2013, 288, 7996–8003. [Google Scholar] [CrossRef]
- Vigoda, M.B.; Argaman, L.; Kournos, M.; Margalit, H. Unraveling the interplay between a small RNA and RNase E in bacteria. Nucleic Acids Res. 2024, 52, 8947–8966. [Google Scholar] [CrossRef]
- Folichon, M.; Arluison, V.; Pellegrini, O.; Huntzinger, E.; Regnier, P.; Hajnsdorf, E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003, 31, 7302–7310. [Google Scholar] [CrossRef]
- Moll, I.; Afonyushkin, T.; Vytvytska, O.; Kaberdin, V.R.; Blasi, U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA 2003, 9, 1308–1314. [Google Scholar] [CrossRef]
- Kornienko, I.V.; Aramova, O.Y.; Tishchenko, A.A.; Rudoy, D.V.; Chikindas, M.L. RNA Stability: A Review of the Role of Structural Features and Environmental Conditions. Molecules 2024, 29, 5978. [Google Scholar] [CrossRef]
- Sorger-Domenigg, T.; Sonnleitner, E.; Kaberdin, V.R.; Blasi, U. Distinct and overlapping binding sites of Pseudomonas aeruginosa Hfq and RsmA proteins on the non-coding RNA RsmY. Biochem. Biophys. Res. Commun. 2007, 352, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Wien, F.; Gragera, M.; Matsuo, T.; Moroy, G.; Bueno-Carrasco, M.T.; Arranz, R.; Cossa, A.; Martel, A.; Bordallo, H.N.; Rudic, S.; et al. Amyloid-like DNA bridging: A new mode of DNA shaping. Nucleic Acids Res. 2025, 53, gkaf169. [Google Scholar] [CrossRef]
- Cossa, A.; Trepout, S.; Wien, F.; Groen, J.; Le Brun, E.; Turbant, F.; Besse, L.; Pereiro, E.; Arluison, V. Cryo soft X-ray tomography to explore Escherichia coli nucleoid remodeling by Hfq master regulator. J. Struct. Biol. 2022, 214, 107912. [Google Scholar] [CrossRef]
- Cech, G.M.; Pakula, B.; Kamrowska, D.; Wegrzyn, G.; Arluison, V.; Szalewska-Palasz, A. Hfq protein deficiency in Escherichia coli affects ColE1-like but not lambda plasmid DNA replication. Plasmid 2014, 73, 10–15. [Google Scholar] [CrossRef]
- Kubiak, K.; Wien, F.; Yadav, I.; Jones, N.C.; Vrønning Hoffmann, S.; Le Cam, E.; Cossa, A.; Geinguenaud, F.; van der Maarel, J.R.C.; Węgrzyn, G.; et al. Amyloid-like Hfq interaction with single-stranded DNA: Involvement in recombination and replication in Escherichia coli. QRB Discovery 2022, 3, e15. [Google Scholar] [CrossRef]
- Parekh, V.J.; Niccum, B.A.; Shah, R.; Rivera, M.A.; Novak, M.J.; Geinguenaud, F.; Wien, F.; Arluison, V.; Sinden, R.R. Role of Hfq in Genome Evolution: Instability of G-Quadruplex Sequences in E. coli. Microorganisms 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gottesman, S. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes. Dev. 2017, 31, 1382–1395. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.J.; Wilusz, J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013, 10, 592–601. [Google Scholar] [CrossRef]
- Mura, C.; Randolph, P.S.; Patterson, J.; Cozen, A.E. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013, 10, 636–651. [Google Scholar] [CrossRef]
- Brennan, R.G.; Link, T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007, 10, 125–133. [Google Scholar] [CrossRef]
- Arluison, V.; Folichon, M.; Marco, S.; Derreumaux, P.; Pellegrini, O.; Seguin, J.; Hajnsdorf, E.; Regnier, P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004, 271, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.S.; Moller-Jensen, J.; Brennan, R.G.; Valentin-Hansen, P. C-Terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 2010, 404, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Fortas, E.; Piccirilli, F.; Malabirade, A.; Militello, V.; Trepout, S.; Marco, S.; Taghbalout, A.; Arluison, V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015, 35, e00190. [Google Scholar] [CrossRef]
- Berbon, M.; Martinez, D.; Morvan, E.; Grelard, A.; Kauffmann, B.; Waeytens, J.; Wien, F.; Arluison, V.; Habenstein, B. Hfq C-terminal region forms a beta-rich amyloid-like motif without perturbing the N-terminal Sm-like structure. Commun. Biol. 2023, 6, 1075. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.; Debelouchina, G.T.; Bayro, M.J.; Clare, D.K.; Caporini, M.A.; Bajaj, V.S.; Jaroniec, C.P.; Wang, L.; Ladizhansky, V.; Muller, S.A.; et al. Atomic structure and hierarchical assembly of a cross-beta amyloid fibril. Proc. Natl. Acad. Sci. USA 2013, 110, 5468–5473. [Google Scholar] [CrossRef]
- Gremer, L.; Scholzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril structure of amyloid-beta(1-42) by cryo-electron microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef]
- Ju, Y.; Tam, K.Y. Pathological mechanisms and therapeutic strategies for Alzheimer’s disease. Neural Regen. Res. 2022, 17, 543–549. [Google Scholar] [CrossRef]
- Maury, C.P. The emerging concept of functional amyloid. J. Intern. Med. 2009, 265, 329–334. [Google Scholar] [CrossRef]
- Otzen, D.; Riek, R. Functional Amyloids. Cold Spring Harb. Perspect. Biol. 2019, 11, a033860. [Google Scholar] [CrossRef]
- Giraldo, R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc. Natl. Acad. Sci. USA 2007, 104, 17388–17393. [Google Scholar] [CrossRef]
- Khambhati, K.; Patel, J.; Saxena, V.; A, P.; Jain, N. Gene Regulation of Biofilm-Associated Functional Amyloids. Pathogens 2021, 10, 490. [Google Scholar] [CrossRef] [PubMed]
- Vecerek, B.; Rajkowitsch, L.; Sonnleitner, E.; Schroeder, R.; Blasi, U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008, 36, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Vincent, H.A.; Henderson, C.A.; Stone, C.M.; Cary, P.D.; Gowers, D.M.; Sobott, F.; Taylor, J.E.; Callaghan, A.J. The low-resolution solution structure of Vibrio cholerae Hfq in complex with Qrr1 sRNA. Nucleic Acids Res. 2012, 40, 8698–8710. [Google Scholar] [CrossRef] [PubMed]
- Salim, N.N.; Faner, M.A.; Philip, J.A.; Feig, A.L. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012, 40, 8021–8032. [Google Scholar] [CrossRef]
- Turbant, F.; Wu, P.; Wien, F.; Arluison, V. The Amyloid Region of Hfq Riboregulator Promotes DsrA:rpoS RNAs Annealing. Biology 2021, 10, 900. [Google Scholar] [CrossRef]
- Kavita, K.; Zhang, A.; Tai, C.H.; Majdalani, N.; Storz, G.; Gottesman, S. Multiple in vivo roles for the C-terminal domain of the RNA chaperone Hfq. Nucleic Acids Res. 2022, 50, 1718–1733. [Google Scholar] [CrossRef]
- Sarni, S.H.; Roca, J.; Du, C.; Jia, M.; Li, H.; Damjanovic, A.; Malecka, E.M.; Wysocki, V.H.; Woodson, S.A. Intrinsically disordered interaction network in an RNA chaperone revealed by native mass spectrometry. Proc. Natl. Acad. Sci. USA 2022, 119, e2208780119. [Google Scholar] [CrossRef]
- Malabirade, A.; Morgado-Brajones, J.; Trepout, S.; Wien, F.; Marquez, I.; Seguin, J.; Marco, S.; Velez, M.; Arluison, V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017, 7, 10724. [Google Scholar] [CrossRef]
- Turbant, F.; Waeytens, J.; Campidelli, C.; Bombled, M.; Martinez, D.; Grelard, A.; Habenstein, B.; Raussens, V.; Velez, M.; Wien, F.; et al. Unraveling Membrane Perturbations Caused by the Bacterial Riboregulator Hfq. Int. J. Mol. Sci. 2022, 23, 8739. [Google Scholar] [CrossRef]
- Diestra, E.; Cayrol, B.; Arluison, V.; Risco, C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS ONE 2009, 4, e8301. [Google Scholar] [CrossRef]
- Taghbalout, A.; Yang, Q.; Arluison, V. The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem. J. 2014, 458, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, Y.; Yagi, M.; Morita, T.; Aiba, H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 2011, 79, 419–432. [Google Scholar] [CrossRef]
- Obregon, K.A.; Hoch, C.T.; Sukhodolets, M.V. Sm-like protein Hfq: Composition of the native complex, modifications, and interactions. Biochim. Biophys. Acta 2015, 1854, 950–966. [Google Scholar] [CrossRef]
- Qiang, W.; Yau, W.M.; Schulte, J. Fibrillation of beta amyloid peptides in the presence of phospholipid bilayers and the consequent membrane disruption. Biochim. Biophys. Acta 2015, 1848, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Nutini, A. Amyloid oligomers and their membrane toxicity—A perspective study. Prog. Biophys. Mol. Biol. 2024, 187, 9–20. [Google Scholar] [CrossRef]
- Turbant, F.; Machiels, Q.; Waeytens, J.; Wien, F.; Arluison, V. The Amyloid Assembly of the Bacterial Hfq Is Lipid-Driven and Lipid-Specific. Int. J. Mol. Sci. 2024, 25, 1434. [Google Scholar] [CrossRef]
- Graham, L.L.; Harris, R.; Villiger, W.; Beveridge, T.J. Freeze-substitution of gram-negative eubacteria: General cell morphology and envelope profiles. J. Bacteriol. 1991, 173, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Sartorio, M.G.; Pardue, E.J.; Feldman, M.F.; Haurat, M.F. Bacterial Outer Membrane Vesicles: From Discovery to Applications. Annu. Rev. Microbiol. 2021, 75, 609–630. [Google Scholar] [CrossRef]
- Kulp, A.; Kuehn, M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef]
- Furuyama, N.; Sircili, M.P. Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. Biomed. Res. Int. 2021, 2021, 1490732. [Google Scholar] [CrossRef]
- Tsatsaronis, J.A.; Franch-Arroyo, S.; Resch, U.; Charpentier, E. Extracellular Vesicle RNA: A Universal Mediator of Microbial Communication? Trends Microbiol. 2018, 26, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Microbiol. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Blenkiron, C.; Simonov, D.; Muthukaruppan, A.; Tsai, P.; Dauros, P.; Green, S.; Hong, J.; Print, C.G.; Swift, S.; Phillips, A.R. Uropathogenic Escherichia coli Releases Extracellular Vesicles That Are Associated with RNA. PLoS ONE 2016, 11, e0160440. [Google Scholar] [CrossRef]
- Lee, H.J. Microbe-Host Communication by Small RNAs in Extracellular Vesicles: Vehicles for Transkingdom RNA Transportation. Int. J. Mol. Sci. 2019, 20, 1487. [Google Scholar] [CrossRef]
- Dauros-Singorenko, P.; Blenkiron, C.; Phillips, A.; Swift, S. The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiol. Lett. 2018, 365, fny023. [Google Scholar] [CrossRef]
- Ghosal, A.; Upadhyaya, B.B.; Fritz, J.V.; Heintz-Buschart, A.; Desai, M.S.; Yusuf, D.; Huang, D.; Baumuratov, A.; Wang, K.; Galas, D.; et al. The extracellular RNA complement of Escherichia coli. Microbiologyopen 2015, 4, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Koeppen, K.; Hampton, T.H.; Jarek, M.; Scharfe, M.; Gerber, S.A.; Mielcarz, D.W.; Demers, E.G.; Dolben, E.L.; Hammond, J.H.; Hogan, D.A.; et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 2016, 12, e1005672. [Google Scholar] [CrossRef]
- Malabirade, A.; Habier, J.; Heintz-Buschart, A.; May, P.; Godet, J.; Halder, R.; Etheridge, A.; Galas, D.; Wilmes, P.; Fritz, J.V. The RNA Complement of Outer Membrane Vesicles from Salmonella enterica Serovar Typhimurium Under Distinct Culture Conditions. Front. Microbiol. 2018, 9, 2015. [Google Scholar] [CrossRef] [PubMed]
- Diallo, I.; Ho, J.; Lambert, M.; Benmoussa, A.; Husseini, Z.; Lalaouna, D.; Masse, E.; Provost, P. A tRNA-derived fragment present in E. coli OMVs regulates host cell gene expression and proliferation. PLoS Pathog. 2022, 18, e1010827. [Google Scholar] [CrossRef]
- Turbant, F.; Waeytens, J.; Blache, A.; Esnouf, E.; Raussens, V.; Wegrzyn, G.; Achouak, W.; Wien, F.; Arluison, V. Interactions and Insertion of Escherichia coli Hfq into Outer Membrane Vesicles as Revealed by Infrared and Orientated Circular Dichroism Spectroscopies. Int. J. Mol. Sci. 2023, 24, 11424. [Google Scholar] [CrossRef]
- Ajam-Hosseini, M.; Akhoondi, F.; Parvini, F.; Fahimi, H. Gram-negative bacterial sRNAs encapsulated in OMVs: An emerging class of therapeutic targets in diseases. Front. Cell Infect. Microbiol. 2023, 13, 1305510. [Google Scholar] [CrossRef]
- Turbant, F.; El Hamoui, O.; Partouche, D.; Sandt, C.; Busi, F.; Wien, F.; Arluison, V. Identification and characterization of the Hfq bacterial amyloid region DNA interactions. BBA Adv. 2021, 1, 100029. [Google Scholar] [CrossRef] [PubMed]
- Sledjeski, D.; Gottesman, S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 2003–2007. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Liu, X.; Yang, L.; Sun, Y.; Gong, Q.; Wu, J.; Shi, Y. The important conformational plasticity of DsrA sRNA for adapting multiple target regulation. Nucleic Acids Res. 2017, 45, 9625–9639. [Google Scholar] [CrossRef]
- Oliver, P.M.; Crooks, J.A.; Leidl, M.; Yoon, E.J.; Saghatelian, A.; Weibel, D.B. Localization of anionic phospholipids in Escherichia coli cells. J. Bacteriol. 2014, 196, 3386–3398. [Google Scholar] [CrossRef]
- Cronan, J.E., Jr. Phospholipid alterations during growth of Escherichia coli. J. Bacteriol. 1968, 95, 2054–2061. [Google Scholar] [CrossRef] [PubMed]
- Kucerka, N.; Heberle, F.A.; Pan, J.; Katsaras, J. Structural Significance of Lipid Diversity as Studied by Small Angle Neutron and X-ray Scattering. Membranes 2015, 5, 454–472. [Google Scholar] [CrossRef]
- Mileykovskaya, E.; Dowhan, W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta 2009, 1788, 2084–2091. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Huang, K.C.; Wingreen, N.S. Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys. J. 2008, 95, 1034–1049. [Google Scholar] [CrossRef]
- Lopez, D.; Kolter, R. Functional microdomains in bacterial membranes. Genes. Dev. 2010, 24, 1893–1902. [Google Scholar] [CrossRef]
- Lopez, D. Molecular composition of functional microdomains in bacterial membranes. Chem. Phys. Lipids 2015, 192, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Wessel, A.K.; Yoshii, Y.; Reder, A.; Boudjemaa, R.; Szczesna, M.; Betton, J.M.; Bernal-Bayard, J.; Beloin, C.; Lopez, D.; Volker, U.; et al. Escherichia coli SPFH Membrane Microdomain Proteins HflKC Contribute to Aminoglycoside and Oxidative Stress Tolerance. Microbiol. Spectr. 2023, 11, e0176723. [Google Scholar] [CrossRef] [PubMed]
- Domenech, O.; Sanz, F.; Montero, M.T.; Hernandez-Borrell, J. Thermodynamic and structural study of the main phospholipid components comprising the mitochondrial inner membrane. Biochim. Biophys. Acta 2006, 1758, 213–221. [Google Scholar] [CrossRef]
- Schlame, M. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. J. Lipid Res. 2008, 49, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Card, G.L.; Trautman, J.K. Role of anionic lipid in bacterial membranes. Biochim. Biophys. Acta 1990, 1047, 77–82. [Google Scholar] [CrossRef]
- Pramanik, J.; Keasling, J.D. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 1997, 56, 398–421. [Google Scholar] [CrossRef]
- Ahmadi Badi, S.; Bruno, S.P.; Moshiri, A.; Tarashi, S.; Siadat, S.D.; Masotti, A. Small RNAs in Outer Membrane Vesicles and Their Function in Host-Microbe Interactions. Front. Microbiol. 2020, 11, 1209. [Google Scholar] [CrossRef] [PubMed]
- Hussein, M.; Jasim, R.; Gocol, H.; Baker, M.; Thombare, V.J.; Ziogas, J.; Purohit, A.; Rao, G.G.; Li, J.; Velkov, T. Comparative Proteomics of Outer Membrane Vesicles from Polymyxin-Susceptible and Extremely Drug-Resistant Klebsiella pneumoniae. mSphere 2023, 8, e0053722. [Google Scholar] [CrossRef]
- Zhu, L.Q.; Gangopadhyay, T.; Padmanabha, K.P.; Deutscher, M.P. Escherichia coli rna gene encoding RNase I: Cloning, overexpression, subcellular distribution of the enzyme, and use of an rna deletion to identify additional RNases. J. Bacteriol. 1990, 172, 3146–3151. [Google Scholar] [CrossRef]
- Beales, P.A.; Bergstrom, C.L.; Geerts, N.; Groves, J.T.; Vanderlick, T.K. Single vesicle observations of the cardiolipin-cytochrome C interaction: Induction of membrane morphology changes. Langmuir 2011, 27, 6107–6115. [Google Scholar] [CrossRef]
- Beltran-Heredia, E.; Tsai, F.C.; Salinas-Almaguer, S.; Cao, F.J.; Bassereau, P.; Monroy, F. Membrane curvature induces cardiolipin sorting. Commun. Biol. 2019, 2, 225. [Google Scholar] [CrossRef] [PubMed]
- Renner, L.D.; Weibel, D.B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. USA 2011, 108, 6264–6269. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Kantor, G.D.; Nishijima, M.; Newman, K.F. Cardiolipin accumulation in the inner and outer membranes of Escherichia coli mutants defective in phosphatidylserine synthetase. J. Bacteriol. 1979, 139, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Ghio, S.; Camilleri, A.; Caruana, M.; Ruf, V.C.; Schmidt, F.; Leonov, A.; Ryazanov, S.; Griesinger, C.; Cauchi, R.J.; Kamp, F.; et al. Cardiolipin Promotes Pore-Forming Activity of Alpha-Synuclein Oligomers in Mitochondrial Membranes. ACS Chem. Neurosci. 2019, 10, 3815–3829. [Google Scholar] [CrossRef]
- Rocha-Roa, C.; Orjuela, J.D.; Leidy, C.; Cossio, P.; Aponte-Santamaria, C. Cardiolipin prevents pore formation in phosphatidylglycerol bacterial membrane models. FEBS Lett. 2021, 595, 2701–2714. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Regulated Control of the Assembly and Diversity of LPS by Noncoding sRNAs. Biomed. Res. Int. 2015, 2015, 153561. [Google Scholar] [CrossRef]
- Tan, B.K.; Bogdanov, M.; Zhao, J.; Dowhan, W.; Raetz, C.R.; Guan, Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc. Natl. Acad. Sci. USA 2012, 109, 16504–16509. [Google Scholar] [CrossRef]
- Guisbert, E.; Rhodius, V.A.; Ahuja, N.; Witkin, E.; Gross, C.A. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J. Bacteriol. 2007, 189, 1963–1973. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velez, M.; Arluison, V. Does the Hfq Protein Contribute to RNA Cargo Translocation into Bacterial Outer Membrane Vesicles? Pathogens 2025, 14, 399. https://doi.org/10.3390/pathogens14040399
Velez M, Arluison V. Does the Hfq Protein Contribute to RNA Cargo Translocation into Bacterial Outer Membrane Vesicles? Pathogens. 2025; 14(4):399. https://doi.org/10.3390/pathogens14040399
Chicago/Turabian StyleVelez, Marisela, and Véronique Arluison. 2025. "Does the Hfq Protein Contribute to RNA Cargo Translocation into Bacterial Outer Membrane Vesicles?" Pathogens 14, no. 4: 399. https://doi.org/10.3390/pathogens14040399
APA StyleVelez, M., & Arluison, V. (2025). Does the Hfq Protein Contribute to RNA Cargo Translocation into Bacterial Outer Membrane Vesicles? Pathogens, 14(4), 399. https://doi.org/10.3390/pathogens14040399




