Antibiotic-Free Selection in Biotherapeutics: Now and Forever
Abstract
:1. Introduction
Regulatory Concern
- -
- It is strongly advised to avoid or minimize the use of any kind of antibiotics in cell or bacterial culture.
- -
- If antibiotics are nevertheless used, it is mandatory to minimize their amount and to monitor the presence of traces in the final product.
- -
- The rationale for the use of antibiotics must be clearly documented in the Common Technical Document (CTD).
- -
- Penicillin, and more generally β-lactams and streptomycin, must not be used in reason of potential concerns with hyper reactivity of some patients to antibiotics of the β-lactam family
- -
- Kanamycin and neomycin are the preferred choice and still tolerated.
- -
- The use of antibiotic resistance markers is generally discouraged, and if used, the in vivo outcome and effect needs to be evaluated.
2. Vector Stabilization
Multimer Resolution
3. Antibiotic-Free Selection and Plasmid Maintenance
| Strain | Strain Modification | Plasmid Maintenance Gene | Antibiotic Resistance Gene Used for Selection | Application | Reference |
|---|---|---|---|---|---|
| Complementation of an essential gene | |||||
| Escherichia coli | dapD mutation → DAP auxotrohpy | dapD | No | DNA and protein expression | Degryse, [32] |
| Salmonella typhimurium | Δasd → DAP auxotrophy | asd | No | Live vaccine | Galan et al. [33] |
| Salmonella typhi | Δasd → DAP auxotrophy | asd | No | Vaccin delivery | Tacket et al. [34] |
| Salmonella typhimurium and S.typhi | thyA mutation → Thymidine auxotrophy | ThyA | No | Live vaccine | Morona et al. [35] |
| Vibrio cholerae | glnA with an internal deletion → Glutamine auxotrophy | glnA | No but presence of TetR and CmR genes on plasmid | Vaccine delivery | Ryan et al. [36] |
| Salmonella enterica serovar Choleraesuis | Δasd → DAP auxotrophy | asd | No | Live vaccine expressing somatostatin | Liang, et al. [6] |
| Escherichia coli | ΔProBA → Proline auxotrophy | proBA | Yes, CmR | protein expression | Fiedler et al. [37] |
| Escherichia coli | ΔTpiA →no growth on glycerol | TpiA | No but presence of AmpR and KanaR genes on plasmid | protein expression | Velur Selvamani, et al. [38] |
| Escherichia coli | ArgE amber mutation + pir116 → arginine auxotrophy | Phe sup tRNA + ori γ | No | DNA (pCOR) | Soubrier et al. [39] |
| Escherichia coli | ThyA amber mutation → thymidine auxotrophy | His sup tRNA | No | DNA (pFAR) | Marie, et al. [40] |
| Salmonella enteritidis | Δasd → DAP auxotrophy | asd | No | Ghost vaccine, live vaccine | Jawale lee, [41] |
| Escherichia coli | ΔglyA → Glycine auxotrohpy | glyA | protein expression | Vidal et al. [42] | |
| Escherichia coli | ΔQAPRTase → NAD auxotrophy | QPARTase | protein expression | Dong, et al. [43] | |
| Bacillus amyloliquefaciens | ΔgspA → G3P auxotrophy | glycerine-3-phosphate dehydrogenase | No | protein expression | US2010/0248306A1 |
| Escherichia coli | ΔaraD | araD | No | protein expression | US2007/0036822 |
| Post Segregational Killing | |||||
| Escherichia coli | - | hok/sok (parB locus) | Yes, bla | DNA and protein expression | Gerdes et al. [44] |
| Escherichia coli | - | hok/sok, parDE | Yes, KanR, AmpR | DNA and protein expression | Pecota et al. [45] |
| Escherichia coli | - | hok/sok (parB locus) | DNA and protein expression | Schweder et al. [46] | |
| Salmonella typhi | - | hok/sok and parA | Yes, bla | Live vaccine | Galen et al. [47] |
| \Escherichia coli | - | ccdA/ccdB | Yes, AmpR | protein expression | Wegerer, et al. [48] |
| Escherichia coli | ccdB gene insertion | ccdA | No but AmpR or spectinomycinR gene present on plasmid | DNA and protein expression | Szpirer et al. [49]. |
| Streptomyces lividans | YoeBsl gene | YefMsl gene | No, but NeoR gene present on plasmid | protein production | Sevilliano et al. [50] |
| Chinese Ovary Hamster cell line (CHO) | Kid | Kis | No but G418R gene present on plasmid | protein production | Nehlsen, et al. [51] |
| Repression of toxic gene | |||||
| Escherichia coli | cI-sacB | cI repressor | No but CmR gene present on plasmid | DNA production | WO2010/135742 |
| Non-antibiotic resistance | |||||
| Pseudomonas putida | Rifr Cmr hsdR1, prototrophic | tellurite resistance gene | RifR and CmR strain but no resistance on plasmid | Bioconversion of toluene | Sanchez-romero [52] |
| Escherichia coli | - | Fabl | no | DNA and protein production | Goh and Good [53] |
| Pseudomonas putida/Klebsiella pneumoniae | - | Biophalos (herbicide), mercury or arsenic resistance gene | No but KanR gene present on plasmid | Strain engineering | Herrero et al. [54] |
| RNA-based drugless systems | |||||
| Escherichia coli | murA under control of TetR (including a RNAI complementary seq.) | RNAi (ColE1 ori) | no | DNA or protein production | Pfaffenzeller et al. [55] |
| Escherichia coli | murA under control of TetR (including a RNAI complementary seq.) | RNAi (ColE1 ori) | no | DNA production | Mairhofer et al. [56] |
| Escherichia coli | RNA-IN sacB | RNA-OUT | no | DNA production | Luke, et al. [57] |
| Operator Repressor Titration systems | |||||
| Escherichia coli | lac-DapD | lacO | no | DNA or protein production | Nuttall, et al. [58], Cranenburgh, et al. [59] |
| Salmonella enterica serovar Typhimurium | lac-DapD | lacO | no | Live vaccine expressing Yersinia pestis F1 antigen | Garmory, et al. [60] |
| Chromosomal integration (plasmid free) | |||||
| Bacillus subtilis | Coat gene in fusion with heterologous peptide | No plasmid | - | Protein display on spores | Iwanicki et al. [61] |
| Escherichia coli | heterologous gene to be expressed | No plasmid | - | protein expression | Striedner, et al. [62] |
| Escherichia coli | GDP-l-fucose de novo pathway genes and H. pylori futC | No plasmid | - | protein production by metabolism modification | Baumgartner et al. [64] |
| Minicircles | |||||
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Chen et al. [65] |
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Chang, et al. [66] |
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Huang, et al. [67] |
| Escherichia coli | ΔendA + PhiC31 integrase gene multiple insertion | Kan resistance gene in producer plasmid | Yes, KanR | DNA production for gene therapy | Kay, et al. [68] |
| Escherichia coli | ΔendA + PhiC31 integrase gene multiple insertion | Kan resistance gene in producer plasmid | Yes, KanR | DNA production for gene therapy | Yi, et al. [69] |
| Escherichia coli | - | Amp resistance gene in producer plasmid | Yes, AmpR | DNA production for gene therapy | Zhang, et al. [70] |





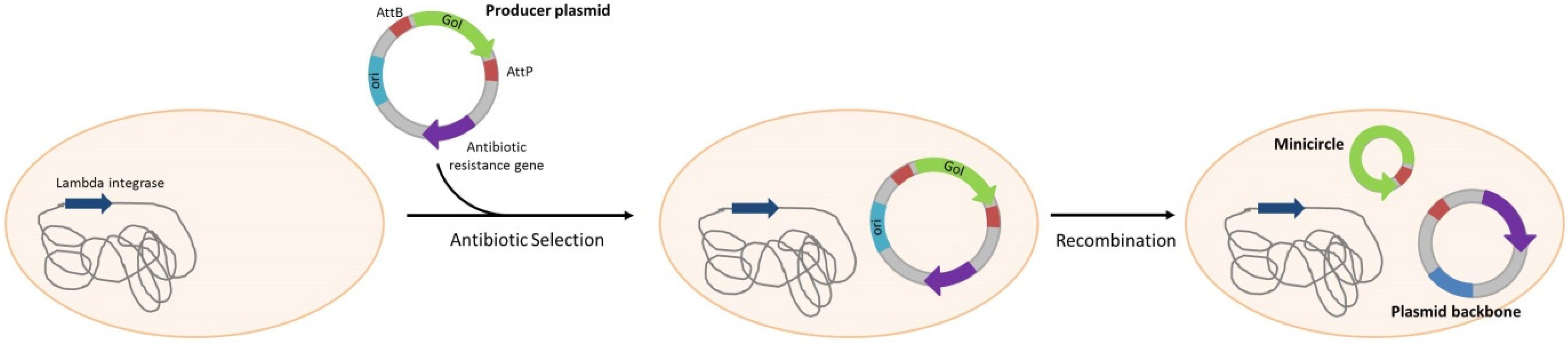
4. Gene Therapy; DNA Immunization
4.1. DNA Vaccines for Human Application
4.2. Use of Viral Vectors
5. Live Vaccine; Delivery Systems
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Livermore, D.M. Introduction: The challenge of multiresistance. Int. J. Antimicrob. Agents 2007, 29 (Suppl. S3), S1–7. [Google Scholar] [CrossRef] [PubMed]
- Bax, R.; Green, S. Antibiotics: The changing regulatory and pharmaceutical industry paradigm. J. Antimicrobi. Chemother. 2015. [Google Scholar] [CrossRef]
- Finch, R.; Hunter, P.A. Antibiotic resistance—Action to promote new technologies: Report of an EU Intergovernmental Conference held in Birmingham, UK, 12–13 December 2005. J. Antimicrob. Chemother. 2006, 58 (Suppl. S1), i3–i22. [Google Scholar] [CrossRef] [PubMed]
- Bakker, J.M.; Bleeker, W.K.; Parren, P.W. Therapeutic antibody gene transfer: An active approach to passive immunity. Mol Ther. 2004, 10, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Armengol, G.; Ruiz, L.M.; Orduz, S. The injection of plasmid DNA in mouse muscle results in lifelong persistence of DNA, gene expression, and humoral response. Mol. biotechnol. 2004, 27, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Liang, A.; Riaz, H.; Dong, F.; Luo, X.; Yu, X.; Han, Y.; Chong, Z.; Han, L.; Guo, A.; Yang, L. Evaluation of efficacy, biodistribution and safety of antibiotic-free plasmid encoding somatostatin genes delivered by attenuated Salmonella enterica serovar Choleraesuis. Vaccine 2014, 32, 1368–1374. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.J.; Bax, R.P. Challenges in developing new antibacterial drugs. Curr. Opin. Investig. Drugs 2009, 10, 157–163. [Google Scholar] [PubMed]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Tuller, T. Codon bias, tRNA pools and horizontal gene transfer. Mob. Genet. elements. 2011, 1, 75–77. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M. Horizontal gene transfer in human pathogens. Crit. rev. Microbiol. 2015, 41, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Datta, N.; Kontomichalou, P. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 1965, 208, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Garber, N.; Friedman, J. Beta-lactamase and the resistance of Pseudomonas aeruginosa to various penicillins and cephalosporins. J. Gen. Microbiol. 1970, 64, 343–352. [Google Scholar] [CrossRef]
- Wiedenbeck, J.; Cohan, F.M. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 2011, 35, 957–976. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Betterton, M.D.; Knight, R. Using the nucleotide substitution rate matrix to detect horizontal gene transfer. BMC bioinform. 2006, 7, 476. [Google Scholar] [CrossRef]
- Roberts, A.P.; Kreth, J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front. Cell. Infect. Microbiol. 2014, 4, 124. [Google Scholar] [CrossRef] [PubMed]
- Sodoyer Regis, C.V.; Peubez, I.; Mignon, C. Antibiotic-free selection for: Moving towards a new “gold standard”. In Antibiotic Resistant Bacteria—A Continuous Challenge in the New Millennium; Marina, P., Ed.; InTech: Rijeka, Croatia, 2012; pp. 531–548. [Google Scholar]
- Georghiou, S.B.; Magana, M.; Garfein, R.S.; Catanzaro, D.G.; Catanzaro, A.; Rodwell, T.C. Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: A systematic review. PLoS ONE 2012, 7, e33275. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Hiraga, S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 1983, 80, 4784–4788. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.; Ogura, T.; Hiraga, S. Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 1985, 163, 841–849. [Google Scholar] [PubMed]
- Gerdes, K.; Larsen, J.E.; Molin, S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985, 161, 292–298. [Google Scholar] [PubMed]
- Gerdes, K.; Poulsen, L.K.; Thisted, T.; Nielsen, A.K.; Martinussen, J.; Andreasen, P.H. The hok killer gene family in gram-negative bacteria. New Biol. 1990, 2, 946–956. [Google Scholar] [PubMed]
- Pedersen, K.; Gerdes, K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999, 32, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Steif, A.; Meyer, I.M. he hok mRNA family. RNA biol. 2012, 9, 1399–404. [Google Scholar] [CrossRef] [PubMed]
- Summers, D.K.; Sherratt, D.J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell 1984, 36, 1097–1103. [Google Scholar] [CrossRef] [PubMed]
- Summers, D.K.; Sherratt, D.J. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 1988, 7, 851–858. [Google Scholar] [PubMed]
- Balding, C.; Blaby, I.; Summers, D. A mutational analysis of the ColE1-encoded cell cycle regulator Rcd confirms its role in plasmid stability. Plasmid 2006, 56, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Blaby, I.K.; Summers, D.K. The role of FIS in the Rcd checkpoint and stable maintenance of plasmid ColE1. Microbiology (Reading, England) 2009, 155, 2676–82. [Google Scholar]
- Peubez, I.; Chaudet, N.; Mignon, C.; Hild, G.; Husson, S.; Courtois, V.; de Luca, K.; Speck, D.; Sodoyer, R. Antibiotic-free selection in E. coli: New considerations for optimal design and improved production. Microb. Cell Fact. 2010, 9, 65. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.; Queiroz, J.A.; Domingues, F.C. Evaluating metabolic stress and plasmid stability in plasmid DNA production by Escherichia coli. Biotechnol. Adv. 2012, 30, 691–708. [Google Scholar] [CrossRef] [PubMed]
- Bower, D.M.; Prather, K.L. Engineering of bacterial strains and vectors for the production of plasmid DNA. Appl. Microbiol. Biotechnol. 2009, 82, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Stenler, S.; Blomberg, P.; Smith, C.I. Safety and efficacy of DNA vaccines: Plasmids vs. minicircles. Hum. Vaccin. Immunother. 2014, 10, 1306–1308. [Google Scholar] [CrossRef] [PubMed]
- Degryse, E. Stability of a host-vector system based on complementation of an essential gene in Escherichia coli. J. Biotechnol. 1991, 18, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.E.; Curtiss, R., 3rd. Distribution of the invA. -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: InvA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun. 1991, 59, 2901–2908. [Google Scholar] [PubMed]
- Tacket, C.O.; Kelly, S.M.; Schodel, F.; Losonsky, G.; Nataro, J.P.; Edelman, R.; Levine, M.M.; Curtiss, R., 3rd. Safety and immunogenicity in humans of an attenuated Salmonella typhi vaccine vector strain expressing plasmid-encoded hepatitis B antigens stabilized by the Asd-balanced lethal vector system. Infect. Immun. 1997, 65, 3381–3385. [Google Scholar] [PubMed]
- Morona, R.; Brown, M.H.; Yeadon, J.; Heuzenroeder, M.W.; Manning, P.A. Effect of lipopolysaccharide core synthesis mutations on the production of Vibrio cholerae O-antigen in Escherichia coli K-12. FEMS Microbiol. Lett. 1991, 66, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Ryan, E.T.; Crean, T.I.; Kochi, S.K.; John, M.; Luciano, A.A.; Killeen, K.P.; Klose, K.E.; Calderwood, S.B. Development of a DeltaglnA balanced lethal plasmid system for expression of heterologous antigens by attenuated vaccine vector strains of Vibrio cholerae. Infect. Immun. 2000, 68, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, M.; Skerra, A. proBA complementation of an auxotrophic E. coli strain improves plasmid stability and expression yield during fermenter production of a recombinant antibody fragment. Gene 2001, 274, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Velur Selvamani, R.S.; Telaar, M.; Friehs, K.; Flaschel, E. Antibiotic-free segregational plasmid stabilization in Escherichia coli owing to the knockout of triosephosphate isomerase (tpiA). Microb. Cell Fact. 2014, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Soubrier, F.; Cameron, B.; Manse, B.; Somarriba, S.; Dubertret, C.; Jaslin, G.; Jung, G.; Caer, C.L.; Dang, D.; Mouvault, J.M.; et al. pCOR: A new design of plasmid vectors for nonviral gene therapy. Gene Ther. 1999, 6, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Marie, C.; Vandermeulen, G.; Quiviger, M.; Richard, M.; Preat, V.; Scherman, D. pFARs. plasmids free of antibiotic resistance markers, display high-level transgene expression in muscle, skin and tumour cells. J. Gene Med. 2010, 12, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Jawale, C.V.; Lee, J.H. Development of a biosafety enhanced and immunogenic Salmonella enteritidis ghost using an antibiotic resistance gene free plasmid carrying a bacteriophage lysis system. PLoS ONE 2013, 8, e78193. [Google Scholar] [CrossRef] [PubMed]
- Vidal, L.; Pinsach, J.; Striedner, G.; Caminal, G.; Ferrer, P. Development of an antibiotic-free plasmid selection system based on glycine auxotrophy for recombinant protein overproduction in Escherichia coli. J. Biotechnol. 2008, 134, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.R.; Xiang, L.X.; Shao, J.Z. Novel antibiotic-free plasmid selection system based on complementation of host auxotrophy in the NAD de novo synthesis pathway. Appl. Environ. Microbiol. 2010, 76, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, K.; Bech, F.W.; Jorgensen, S.T.; Lobner-Olesen, A.; Rasmussen, P.B.; Atlung, T.; Boe, L.; Karlstrom, O.; Molin, S.; von Meyenburg, K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986, 5, 2023–2029. [Google Scholar] [PubMed]
- Pecota, D.C.; Kim, C.S.; Wu, K.; Gerdes, K.; Wood, T.K. Combining the hok/sok. parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl. Environ. Microbiol. 1997, 63, 1917–1924. [Google Scholar] [PubMed]
- Schweder, T.; Schmidt, I.; Herrmann, H.; Neubauer, P.; Hecker, M.; Hofmann, K. An expression vector system providing plasmid stability and conditional suicide of plasmid-containing cells. Appl. Environ. Microbiol. 1992, 38, 91–93. [Google Scholar]
- Galen, J.E.; Nair, J.; Wang, J.Y.; Wasserman, S.S.; Tanner, M.K.; Sztein, M.B.; Levine, M.M. Optimization of plasmid maintenance in the attenuated live vector vaccine strain Salmonella typhi CVD 908-htrA. Infect. Immun. 1999, 67, 6424–6433. [Google Scholar] [PubMed]
- Wegerer, A.; Sun, T.; Altenbuchner, J. Optimization of an E. coli l-rhamnose-inducible expression vector: Test of various genetic module combinations. BMC Biotechnol. 2008, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Szpirer, C.Y.; Milinkovitch, M.C. Separate-component-stabilization system for protein and DNA production without the use of antibiotics. Biotechniques 2005, 38, 775–381. [Google Scholar] [CrossRef] [PubMed]
- Sevillano, L.; Diaz, M.; Santamaria, R.I. Stable expression plasmids for Streptomyces based on a toxin-antitoxin system. Microb. Cell Fact. 2013, 12, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nehlsen, K.; Herrmann, S.; Zauers, J.; Hauser, H.; Wirth, D. Toxin-antitoxin based transgene expression in mammalian cells. Nucleic Acids Res. 2010, 38, e32. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Romero, J.M.; Diaz-Orejas, R.; de Lorenzo, V. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 1998, 64, 4040–4046. [Google Scholar] [PubMed]
- Goh, S.; Good, L. Plasmid selection in Escherichia coli using an endogenous essential gene marker. BMC Biotechnol. 2008, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [PubMed]
- Pfaffenzeller, I.; Mairhofer, J.; Striedner, G.; Bayer, K.; Grabherr, R. Using ColE1-derived RNA I for suppression of a bacterially encoded gene: Implication for a novel plasmid addiction system. Biotechnol. J. 2006, 1, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Mairhofer, J.; Pfaffenzeller, I.; Merz, D.; Grabherr, R. A novel antibiotic free plasmid selection system: Advances in safe and efficient DNA therapy. Biotechnol. J. 2008, 3, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.; Carnes, A.E.; Hodgson, C.P.; Williams, J.A. Improved antibiotic-free DNA vaccine vectors utilizing a novel RNA based plasmid selection system. Vaccine 2009, 27, 6454–6459. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, S.D.; Krishnan, U.V.; Hattarki, M.; de Gori, R.; Irving, R.A.; Hudson, P.J. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol. 2001, 38, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Cranenburgh, R.M.; Lewis, K.S.; Hanak, J.A. Effect of plasmid copy number and lac operator sequence on antibiotic-free plasmid selection by operator-repressor titration in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2004, 7, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Garmory, H.S.; Leckenby, M.W.; Griffin, K.F.; Elvin, S.J.; Taylor, R.R.; Hartley, M.G.; Hanak, J.A.; Williamson, E.D.; Cranenburgh, R.M. Antibiotic-free plasmid stabilization by operator-repressor titration for vaccine delivery by using live Salmonella enterica Serovar typhimurium. Infect. Immun. 2005, 73, 2005–2011. [Google Scholar] [CrossRef] [PubMed]
- Iwanicki, A.; Piatek, I.; Stasilojc, M.; Grela, A.; Lega, T.; Obuchowski, M.; Hinc, K. A system of vectors for Bacillus subtilis spore surface display. Microb. Cell Fact. 2014, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Striedner, G.; Pfaffenzeller, I.; Markus, L.; Nemecek, S.; Grabherr, R.; Bayer, K. Plasmid-free T7-based Escherichia coli expression systems. Biotechnol. Bioeng. 2010, 105, 786–794. [Google Scholar] [PubMed]
- Lemuth, K.; Steuer, K.; Albermann, C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb. Cell Fact. 2011, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, F.; Seitz, L.; Sprenger, G.A.; Albermann, C. Construction of Escherichia coli strains with chromosomally integrated expression cassettes for the synthesis of 2'-fucosyllactose. Microb. Cell Fact. 2013, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.Y.; He, C.Y.; Kay, M.A. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005, 16, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Christensen, L.V.; Lee, M.; Kim, S.W. Efficient expression of vascular endothelial growth factor using minicircle DNA for angiogenic gene therapy. J. Control. Release 2008, 125, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Chen, Z.; Hu, S.; Jia, F.; Li, Z.; Hoyt, G.; Robbins, R.C.; Kay, M.A.; Wu, J.C. Novel minicircle vector for gene therapy in murine myocardial infarction. Circulation 2009, 120, S230–S237. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.A.; He, C.Y.; Chen, Z.Y. A robust system for production of minicircle DNA vectors. Nat. Biotechnol. 2010, 28, 1287–1289. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Kim, Y.; Kim, J.; Jung, H.; Rim, Y.A.; Jung, S.M.; Park, S.H.; Ju, J.H. A new strategy to deliver synthetic protein drugs: Self-reproducible biologics using minicircles. Sci. Rep. 2014, 4, 5961. [Google Scholar] [PubMed]
- Zhang, X.; Epperly, M.W.; Kay, M.A.; Chen, Z.Y.; Dixon, T.; Franicola, D.; Greenberger, B.A.; Komanduri, P.; Greenberger, J.S. Radioprotection in vitro and in vivo by minicircle plasmid carrying the human manganese superoxide dismutase transgene. Hum. Gene Ther. 2008, 19, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kong, Q.; Curtiss, R., 3rd. New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb. Pathog. 2013, 58, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.M.; Sali, M.; Leckenby, M.W.; Radford, D.S.; Huynh, H.A.; Delogu, G.; Cranenburgh, R.M.; Cutting, S.M. Oral delivery of a DNA vaccine against tuberculosis using operator-repressor titration in a Salmonella enterica vector. Vaccine 2010, 28, 7523–7528. [Google Scholar] [CrossRef] [PubMed]
- Bloor, A.E.; Cranenburgh, R.M. An efficient method of selectable marker gene excision by Xer recombination for gene replacement in bacterial chromosomes. Appl. Environ. Microbiol. 2006, 72, 2520–2525. [Google Scholar] [CrossRef] [PubMed]
- Raiford, D.W.; Heizer, E.M., Jr.; Miller, R.V.; Doom, T.E.; Raymer, M.L.; Krane, D.E. Metabolic and translational efficiency in microbial organisms. J. Mol. Evol. 2012, 74, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Soubrier, F.; Laborderie, B.; Cameron, B. Improvement of pCOR plasmid copy number for pharmaceutical applications. Appl. Microbiol. Biotechnol. 2005, 66, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Vandermeulen, G.; Marie, C.; Scherman, D.; Preat, V. New generation of plasmid backbones devoid of antibiotic resistance marker for gene therapy trials. Mol. Ther. 2011, 19, 1942–1949. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Harrison, E.M.; Bi, D.; Tai, C.; He, X.; Ou, H.Y.; Rajakumar, K.; Deng, Z. TADB: A web-based resource for Type 2 toxin-antitoxin loci in bacteria and archaea. Nucleic Acids Res. 2011, 39, D606–D611. [Google Scholar] [CrossRef]
- Galen, J.E.; Wang, J.Y.; Chinchilla, M.; Vindurampulle, C.; Vogel, J.E.; Levy, H.; Blackwelder, W.C.; Pasetti, M.F.; Levine, M.M. A new generation of stable. nonantibiotic, low-copy-number plasmids improves immune responses to foreign antigens in Salmonella enterica serovar Typhi live vectors. Infect. Immun. 2010, 78, 337–347. [Google Scholar]
- Stieber, D.; Gabant, P.; Szpirer, C. The art of selective killing: Plasmid toxin/antitoxin systems and their technological applications. Biotechniques 2008, 45, 344–346. [Google Scholar] [CrossRef] [PubMed]
- Diago-Navarro, E.; Hernandez-Arriaga, A.M.; Lopez-Villarejo, J.; Munoz-Gomez, A.J.; Kamphuis, M.B.; Boelens, R.; Lemonnier, M.; Diaz-Orejas, R. parD toxin-antitoxin system of plasmid R1—Basic contributions, biotechnological applications and relationships with closely-related toxin-antitoxin systems. FEBS J. 2010, 277, 3097–3117. [Google Scholar] [CrossRef] [PubMed]
- De la Cueva-Mendez, G.; Mills, A.D.; Clay-Farrace, L.; Diaz-Orejas, R.; Laskey, R.A. Regulatable killing of eukaryotic cells by the prokaryotic proteins Kid and Kis. EMBO J. 2003, 22, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.M.; Carnes, A.E.; Williams, J.A. Development of antibiotic-free selection system for safer DNA vaccination. Methods Mol. Biol. 2014, 1143, 91–111. [Google Scholar] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Tuntufye, H.N.; Goddeeris, B.M. Use of lambda Red-mediated recombineering and Cre/lox for generation of markerless chromosomal deletions in avian pathogenic Escherichia coli. FEMS Microbiol. Lett. 2011, 325, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Bryant, J.A.; Sellars, L.E.; Busby, S.J.; Lee, D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014, 42, 11383–11392. [Google Scholar] [CrossRef] [PubMed]
- Darquet, A.M.; Cameron, B.; Wils, P.; Scherman, D.; Crouzet, J. A new DNA vehicle for nonviral gene delivery: Supercoiled minicircle. Gene Ther. 1997, 4, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Bigger, B.W.; Tolmachov, O.; Collombet, J.M.; Fragkos, M.; Palaszewski, I.; Coutelle, C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem. 2001, 276, 23018–23027. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.S.; Kwon, J.T.; Shim, I.; Kim, H.M.; Kim, P.; Kim, J.C.; Lee, K. Evaluation of toxicity to triclosan in rats following 28days of exposure to aerosol inhalation. Regul. Toxicol. Pharmacol. 2015, 71, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Li, Y.; Zhou, Q.; Xu, Y.; Wang, D. Effect of triclosan on reproduction. DNA damage and heat shock protein gene expression of the earthworm Eisenia fetida. Ecotoxicology 2014, 23, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Lee, M.H.; Joo, H.; Jung, Y.H.; Ahmad, S.; Choi, J.; Lee, S.W. Triclosan resistance in a bacterial fish pathogen aeromonas salmonicida subsp. salmonicida is mediated by an enoyl reductase FabV. J. Microbiol. Biotechnol. 2014. [Google Scholar] [PubMed]
- Oliveira, P.H.; Mairhofer, J. Marker-free plasmids for biotechnological applications—Implications and perspectives. Trends Biotechnol. 2013, 31, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A. Improving DNA vaccine performance through vector design. Curr.Gene Ther. 2014, 14, 170–189. [Google Scholar] [CrossRef] [PubMed]
- Reschner, A.; Scohy, S.; Vandermeulen, G.; Daukandt, M.; Jacques, C.; Michel, B.; Nauwynck, H.; Xhonneux, F.; Preat, V.; Vanderplasschen, A.; et al. Use of Staby((R)) technology for development and production of DNA vaccines free of antibiotic resistance gene. Hum. vaccin. Immunother. 2013, 9, 2203–2210. [Google Scholar] [CrossRef] [PubMed]
- Ginn, S.L.; Alexander, I.E.; Edelstein, M.L.; Abedi, M.R.; Wixon, J. Gene Therapy Clinical Trials Worldwide to 2012. J. Gene Med. 2013, 15, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Note for guidance on the quality, preclinical and clinical aspects of gene transfer medicinal products. Annex on non-clinical testing for indadvertent germline transmission of gene transfer vectors. (2001:648)
- WHO. Guidelines for assuring the quality of non clinical safety evaluation of DNA vaccines. In Proceedings of the 56th Meeting of the WHO Expert Committee on Biological Standardization, Geneva, Switzerland, 24–28 October 2005.
- Beckett, C.G.; Tjaden, J.; Burgess, T.; Danko, J.R.; Tamminga, C.; Simmons, M.; Wu, S.J.; Sun, P.; Kochel, T.; Raviprakash, K.; et al. Evaluation of a prototype dengue-1 DNA vaccine in a Phase 1 clinical trial. Vaccine 2011, 29, 960–968. [Google Scholar] [CrossRef]
- Nikol, S.; Baumgartner, I.; van Belle, E.; Diehm, C.; Visona, A.; Capogrossi, M.C.; Ferreira-Maldent, N.; Gallino, A.; Wyatt, M.G.; Wijesinghe, L.D.; et al. Therapeutic angiogenesis with intramuscular NV1FGF improves amputation-free survival in patients with critical limb ischemia. Mol. Ther. 2008, 16, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Mwau, M.; Cebere, I.; Sutton, J.; Chikoti, P.; Winstone, N.; Wee, E.G.; Beattie, T.; Chen, Y.H.; Dorrell, L.; McShane, H.; et al. A human immunodeficiency virus 1 (HIV-1) clade A vaccine in clinical trials: Stimulation of HIV-specific T-cell responses by DNA and recombinant modified vaccinia virus Ankara (MVA) vaccines in humans. J. Gen. Virol. 2004, 85, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Cranenburgh, R.M.; Hanak, J.A.; Williams, S.G.; Sherratt, D.J. Escherichia coli strains that allow antibiotic-free plasmid selection and maintenance by repressor titration. Nucleic Acids Res. 2001, 29, E26. [Google Scholar] [CrossRef] [PubMed]
- Andries, O.; Kitada, T.; Bodner, K.; Sanders, N.N.; Weiss, R. Synthetic biology devices and circuits for RNA-based “smart vaccines”: A propositional review. Expert Rev. Vaccin. 2015, 14, 313–331. [Google Scholar] [CrossRef]
- Ulmer, J.B.; Mason, P.W.; Geall, A.; Mandl, C.W. RNA-based vaccines. Vaccine 2012, 30, 4414–4418. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, K.; Liljestrom, P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccin. 2015, 14, 177–194. [Google Scholar] [CrossRef]
- Melchiorri, D.; Pani, L.; Gasparini, P.; Cossu, G.; Ancans, J.; Borg, J.J.; Drai, C.; Fiedor, P.; Flory, E.; Hudson, I.; et al. Regulatory evaluation of Glybera in Europe—Two committees, one mission. Nat. Rev. Drug Discov. 2013, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.; Rodriguez, S.; Finlayson, N.; Williams, J.; Carnes, A. Antibiotic-free production of a herpes simplex virus 2 DNA vaccine in a high yield cGMP process. Hum. Vaccin. Immunother. 2013, 9, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Mattia, A.; Merker, R. Regulation of probiotic substances as ingredients in foods: Premarket approval or “generally recognized as safe” notification. Clin. Infect. Dis. 2008, 46, S115–118, discussion S144–151. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Valdez, F.J.; Perez Brandan, C.; Ferreira, A.; Basombrio, M.A. Gene-deleted live-attenuated Trypanosoma cruzi parasites as vaccines to protect against Chagas disease. Expert Rev. Vaccin. 2014, 1–17. [Google Scholar] [CrossRef]
- Trombert, A. Recombinant lactic acid bacteria as delivery vectors of heterologous antigens: The future of vaccination? Benef. Microbes 2014, 1–12. [Google Scholar] [CrossRef]
- Da Costa, A.C.; Nogueira, S.V.; Kipnis, A.; Junqueira-Kipnis, A.P. Recombinant BCG: Innovations on an Old Vaccine. Scope of BCG Strains and Strategies to Improve Long-Lasting Memory. Front. Immunol. 2014, 5, 152. [Google Scholar] [CrossRef] [PubMed]
- Chin’ombe, N.; Ruhanya, V. Recombinant salmonella bacteria vectoring HIV/AIDS vaccines. Open Virol. J. 2013, 7, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.L.; Wang, P.; Tao, H.X.; Liu, X.X.; Wang, Y.C.; Zhan, D.W.; Liu, C.J.; Zhang, Z.S. Removal of antibiotic resistance of live vaccine strain Escherichia coli MM-3 and evaluation of the immunogenicity of the new strain. Acta Biochim. Biophys. Sin. 2006, 38, 844–856. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mignon, C.; Sodoyer, R.; Werle, B. Antibiotic-Free Selection in Biotherapeutics: Now and Forever. Pathogens 2015, 4, 157-181. https://doi.org/10.3390/pathogens4020157
Mignon C, Sodoyer R, Werle B. Antibiotic-Free Selection in Biotherapeutics: Now and Forever. Pathogens. 2015; 4(2):157-181. https://doi.org/10.3390/pathogens4020157
Chicago/Turabian StyleMignon, Charlotte, Régis Sodoyer, and Bettina Werle. 2015. "Antibiotic-Free Selection in Biotherapeutics: Now and Forever" Pathogens 4, no. 2: 157-181. https://doi.org/10.3390/pathogens4020157
APA StyleMignon, C., Sodoyer, R., & Werle, B. (2015). Antibiotic-Free Selection in Biotherapeutics: Now and Forever. Pathogens, 4(2), 157-181. https://doi.org/10.3390/pathogens4020157





