Morphological and Molecular Variation Between Fusarium avenaceum, Fusarium arthrosporioides and Fusarium anguioides Strains
Abstract
:1. Introduction
2. Results
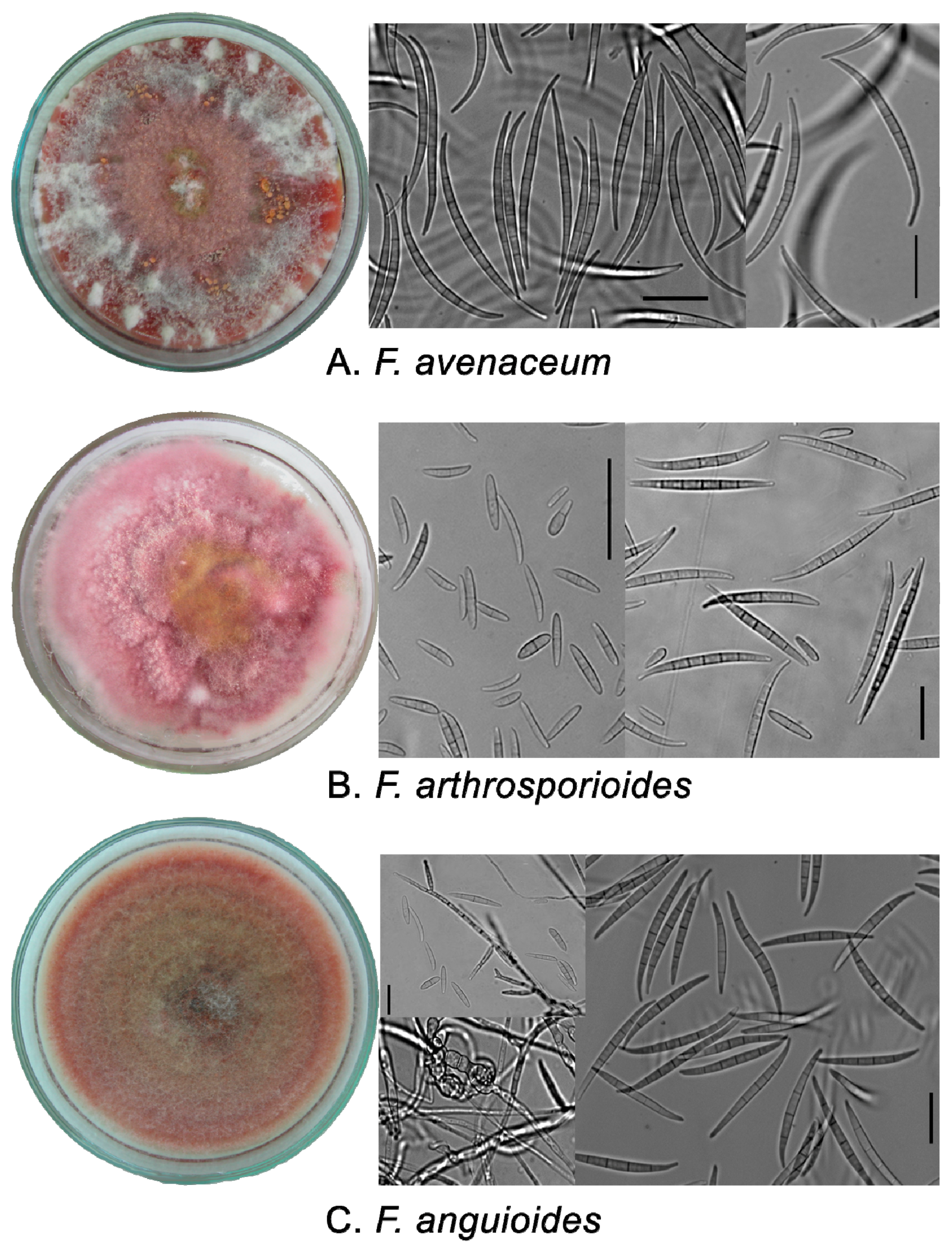
2.1. Phenotypic Characters of Strains
2.2. Pathogenicity of Strains
2.3. Molecular Diversity of Strains
3. Discussion
4. Materials and Methods
4.1. Strains of Fusarium
4.2. Morphological Analyses
4.3. The Analyses of Pathogenicity
4.4. DNA Extraction, PCR and Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Peters, J.C.; Lees, A.K.; Cullen, D.W.; Sullivan, L.; Stroud, G.P.; Cunnington, A.C. Characterization of Fusarium spp. responsible for causing dry rot of potato in Great Britain. Plant Pathol. 2008, 57, 262–271. [Google Scholar] [CrossRef]
- Sørensen, J.L.; Phipps, R.K.; Nielsen, K.F.; Schroers, H.J.; Frank, J.; Thrane, U. Analysis of Fusarium avenaceum metabolites produced during wet apple core rot. J. Agric. Food Chem. 2009, 57, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Sakoda, T.; Yamasaki, N.; Abe, Y.; Yanagisawa, H.; Koike, M. Bulb Rot of Sandersonia aurantiaca caused by Fusarium anguioides and Fusarium sp. Intercepted at Plant Quarantine in Japan. Res. Bull. Plant Prot. Japan 2011, 47, 41–47. [Google Scholar]
- Yli-Mattila, T.; Paavanen-Huhtala, S.; Parikka, P.; Konstantinova, P.; Gagkaeva, T. Molecular and morphological diversity of Fusarium species in Finland and northwestern Russia. Eur. J. Plant Pathol. 2004, 110, 573–585. [Google Scholar] [CrossRef]
- Uhlig, S.; Jestoi, M.; Parikka, P. Fusarium avenaceum—The North European situation. Int J Food Microbiol 2007, 119, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Gräfenhan, T.; Patrick, S.K.; Roscoe, M.; Trelka, R.; Gaba, D.; Chan, J.M.; McKendry, T.; Clear, R.M.; Tittlemier, S.A. Fusarium damage in cereal grains from Western Canada. 1. Phylogenetic Analysis of Moniliformin-Producing Fusarium Species and Their Natural Occurrence in Mycotoxin-Contaminated Wheat, Oats, and Rye. J. Agric. Food Chem. 2013, 61, 5425–5437. [Google Scholar] [CrossRef] [PubMed]
- Hietaniemi, V.; Ramo, S.; Yli-Mattila, T.; Jestoi, M.; Peltonen, S.; Kartio, M.; Sievilainen, E.; Koivisto, T.; Parikka, P. Updated survey of Fusarium species and toxins in Finnish cereal grains. Food. Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2016, 33, 831–848. [Google Scholar] [CrossRef] [PubMed]
- Gavrilova, O.P.; Gannibal, P.B.; Gagkaeva, T.Y. Fusarium and Alternaria fungi in grain of oats grown in the north-western Russia regarding cultivar specificity. Sel’skokhozyaistvennaya biologiya 2016, 51, 111–118. [Google Scholar] [CrossRef]
- Jestoi, M.; Rokka, M.; Yli-Mattila, T.; Parikka, P.; Rizzo, A.; Peltonen, K. Presence and concentrations of the Fusarium-related mycotoxins beauvericin, enniatins and moniliformin in Finnish grain samples. Food Addit. Contam. 2004, 21, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Yli-Mattila, T.; Paavanen-Huhtala, S.; Parikka, P.A.; Jestoi, M.; Klemsdal, S.; Rizzo, A.A. Genetic variation, real-time PCR, metabolites and mycotoxins of Fusarium avenaceum and related species. Mycotox. Res. 2006, 22, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Cook, R.J. Gibberella avenacea sp. n., perfect stage of Fusarium roseum f. sp. cerealis ‘avenaceum’. Phytopathology 1967, 57, 732–736. [Google Scholar]
- Booth, C.; Spooner, B.M. Gibberella avenacea, teleomorph of Fusarium avenaceum, from stems of Pteridium aquilinum. Trans. Br. Mycol. Soc. 1984, 82, 178–180. [Google Scholar] [CrossRef]
- Kerényi, Z.; Moretti, A.; Waalwijk, C.; Oláh, B.; Hornok, L. Mating type sequences in asexually reproducing Fusarium species. Appl. Environ. Microbiol. 2004, 70, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Booth, C. The Genus Fusarium; CommonwealthMycological Institute: Kew, Surrey, UK, 1971; p. 237. [Google Scholar]
- Parikka, P.; Hakala, K.; Tiilikkala, K. Expected shifts in Fusarium species’ composition on cereal grain in Northern Europe due to climate change. Food. Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2012, 29, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Fusarium species: An illustrated manual for identification; Pennsylvania State University Press: University Park, PA, USA, 1983; p. 206. [Google Scholar]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell Publishing: Hoboken, NJ, USA, 2006; p. 388. [Google Scholar]
- Gerlach, W.; Nirenberg, H. The genus Fusarium—A Pictorial Atlas Mitt. Biol. Bundesanst. Ld.; Mitteilungen aus der Biologischen Bundesanstalt fur Land- und Forstwirtschaft Berlin-Dahlem: Berlin, German, 1982; p. 406. [Google Scholar]
- Sherbakoff, C.D. Fusaria of potatoes. In Memoirs of the Cornell University Agricultural Experimental Station; Cornell University. Agricultural Experiment Station: Ithaca, NY, USA, 1915; pp. 87–270. [Google Scholar]
- Geiser, D.M.; Aoki, T.; Bacon, C.W.; Baker, S.E.; Bhattacharyya, M.K.; Brandt, M.E.; Brown, D.W.; Burgess, L.W.; Chulze, S.; Coleman, J.J.; et al. One fungus, one name: defining the genus Fusarium in a scientifically robust way that preserves longstanding use. Phytopathology 2013, 103, 400–408. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Ward, T.J.; Robert, V.A.R.G; Crous, P.W.; Geiser, D.M.; Kang, S. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 2015, 43, 583–595. [Google Scholar] [CrossRef]
- Torbati, M.; Arzanlou, M.; Sandoval-Denis, M.; Crous, P.W. Multigene phylogeny reveals new fungicolous species in the Fusarium tricinctum species complex and novel hosts in the genus Fusarium from Iran. Mycol. Prog. 2018. [Google Scholar] [CrossRef]
- Yli-Mattila, T.; Paavanen-Huhtala, S.; Bulat, S.A.; Alekhina, I.A.; Nirenberg, H.I. Molecular, morphological and phylogenetic analysis of Fusarium avenaceum/F. arthrosporioides/F. tricinctum species complex—A polyphasic approach. Mycol. Res. 2002, 106, 655–669. [Google Scholar] [CrossRef]
- Niessen, L.; Gräfenhan, T.; Vogel, R.F. ATP citrate lyase 1 (asc1) gene-based loop-mediated amplification assay for the detection of the Fusarium tricinctum species complex in pure cultures and in cereal samples. Int. J. Food Microbiol. 2012, 158, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Nalim, F.A.; Elmer, W.H.; McGovern, R.J.; Geiser, D.M. Multilocus phylogenetic diversity of Fusarium avenaceum pathogenic on lisianthus. Phytopathology 2009, 99, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Kulik, T.; Pszczółkowska, A.; Łojko, M. Multilocus phylogenetics show high intraspecific variability within Fusarium avenaceum. Int. J. Mol. Sci. 2011, 12, 5626–5640. [Google Scholar] [CrossRef] [PubMed]
- Stakheev, A.A.; Khairulina, D.R.; Zavriev, S.K. Four-locus phylogeny of Fusarium avenaceum and related species and their species-specific identification based on partial phosphate permease gene sequences. Int. J. Food Microbiol. 2016, 225, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Lysøe, E.; Harris, L.J.; Walkowiak, S.; Subramaniam, R.; Divon, H.H.; Riiser, E.S.; Llorens, C.; Gabaldón, T.; Kistler, H.C.; Jonkers, W.; et al. The genome of the generalist plant pathogen Fusarium avenaceum is enriched with genes involved in redox, signaling and secondary metabolism. PLoS One 2014, 9, e112703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirenberg, H. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Section Liseola. Mitteilungen der Biologischen Bundesanstalt für Land- und Forstwirtschaft 1976, 169, 1–117. [Google Scholar]
- Nelson, P.E.; Toussoun, T.A.; Marasas, W.F.O. Neotypification and emended description of Fusarium anguioides. Mycologia 1995, 8, 543–546. [Google Scholar] [CrossRef]
- Chelkowski, J.; Manka, M. The ability of Fusaria pathogenic to wheat, barley and corn to produce zearalenone. Phytopathol. Z. 1983, 106, 354–359. [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: a guide to methods and applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Turner, A.S.; Lees, A.K.; Rezanoor, H.N.; Nicholson, P. Refinement of PCR-detection of Fusarium avenaceum and evidence from DNA marker studies for phenetic relatedness to Fusarium tricinctum. Plant Pathol. 1998, 47, 278–288. [Google Scholar] [CrossRef]
- Stenglein, S.A.; Balatti, P.A. Genetic diversity of Phaeioisariopsis griseola in Argentina as revealed by pathogenic and molecular markers. Phys. Mol. Plant Pathol. 2006, 68, 158–167. [Google Scholar] [CrossRef]





| No. | Catalogue no.* | Fusarium sp. | Origin | Substrate | Year | Strain no. |
|---|---|---|---|---|---|---|
| 1 | MFG103100 | F. anguioides | Kaliningrad, Russia | oat, grain | 2007 | an35 |
| 2 | MFG109902 | F. anguioides | Vologda, Russia | wheat, grain | 2008 | an36 |
| 3 | MFG112804 | F. anguioides | Novgorod, Russia | barley, grain | 2008 | an37 |
| 4 | MFG115014 | F. anguioides | Kaliningrad, Russia | barley, grain | 2008 | an38 |
| 5 | MFG114003 | F. anguioides | Novgorod, Russia | oat, grain | 2008 | an39 |
| 6 | MFG116504 | F. arthrosporioides | Kirov, Russia | barley, grain | 2008 | ar40 |
| 7 | MFG114605 | F. anguioides | Kaliningrad, Russia | barley, grain | 2008 | an41 |
| 8 | MFG119913 | F. anguioides | Kirov, Russia | oat, grain | 2008 | an42 |
| 9 | MFG118902 | F. anguioides | Kirov, Russia | oat, grain | 2008 | an43 |
| 10 | MFG115015 | F. anguioides | Kaliningrad, Russia | barley, grain | 2008 | an44 |
| 11 | MFG108904 | F. anguioides | Pskov, Russia | barley, grain | 2008 | an45 |
| 12 | MFG58657 | F. anguioides | Poland | air | 2013 | an46 |
| 13 | MFG58314 | F. anguioides | Vladivostok, Russia | rudbeckia, leaves | 2010 | an47 |
| 14 | MFG118702 | F. avenaceum | Pskov, Russia | barley, grain | 2009 | av48 |
| 15 | MFG58656 | F. avenaceum | Poland | air | 2013 | av49 |
| 16 | MFG151200 | F. avenaceum | Leningrad, Russia | oat, grain | 2011 | av50 |
| 17 | MFG58640 | F. avenaceum | Moscow, Russia | wheat, grain | 2004 | av51 |
| 18 | MFG168800 | F. avenaceum | Leningrad, Russia | oat, grain | 2011 | av52 |
| 19 | MFG118401 | F. avenaceum | Pskov, Russia | oat, grain | 2009 | av53 |
| 20 | MFG58426 | F. avenaceum | Krasnodar, Russia | barley, grain | 2011 | av54 |
| 21 | MFG117309 | F. avenaceum | Kirov, Russia | oat, grain | 2008 | av55 |
| 22 | MFG58654 | F. arthrosporioides | Tyumen, Russia | barley, grain | 2013 | ar56 |
| 23 | MFG58655 | F. arthrosporioides | Leningrad, Russia | oat, grain | 2013 | ar57 |
| 24 | BBA64215 | F. arthrosporioides | unknown | unknown | un-known | ar1 |
| 25 | BBA63598 | F. anguioides | Japan | pea | 1928 | an2 |
| 26 | BBA69055 | F. anguioides | Japan | wheat, grain | 1994 | an3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yli-Mattila, T.; Hussien, T.; Gavrilova, O.; Gagkaeva, T. Morphological and Molecular Variation Between Fusarium avenaceum, Fusarium arthrosporioides and Fusarium anguioides Strains. Pathogens 2018, 7, 94. https://doi.org/10.3390/pathogens7040094
Yli-Mattila T, Hussien T, Gavrilova O, Gagkaeva T. Morphological and Molecular Variation Between Fusarium avenaceum, Fusarium arthrosporioides and Fusarium anguioides Strains. Pathogens. 2018; 7(4):94. https://doi.org/10.3390/pathogens7040094
Chicago/Turabian StyleYli-Mattila, Tapani, Taha Hussien, Olga Gavrilova, and Tatiana Gagkaeva. 2018. "Morphological and Molecular Variation Between Fusarium avenaceum, Fusarium arthrosporioides and Fusarium anguioides Strains" Pathogens 7, no. 4: 94. https://doi.org/10.3390/pathogens7040094
APA StyleYli-Mattila, T., Hussien, T., Gavrilova, O., & Gagkaeva, T. (2018). Morphological and Molecular Variation Between Fusarium avenaceum, Fusarium arthrosporioides and Fusarium anguioides Strains. Pathogens, 7(4), 94. https://doi.org/10.3390/pathogens7040094






