In Situ Characterization of Hfq Bacterial Amyloid: A Fourier-Transform Infrared Spectroscopy Study
Abstract
1. Introduction
2. Results and Discussion
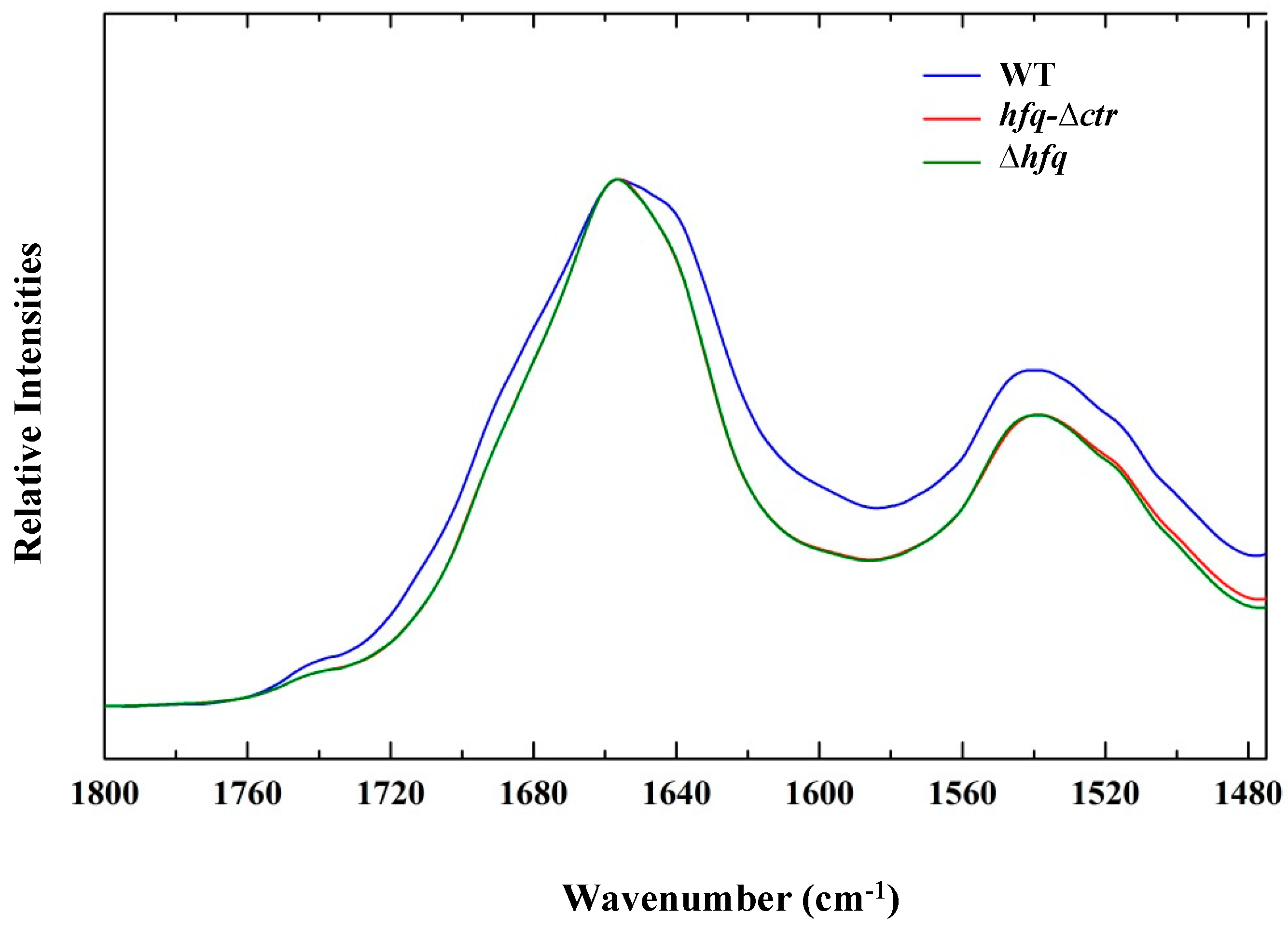
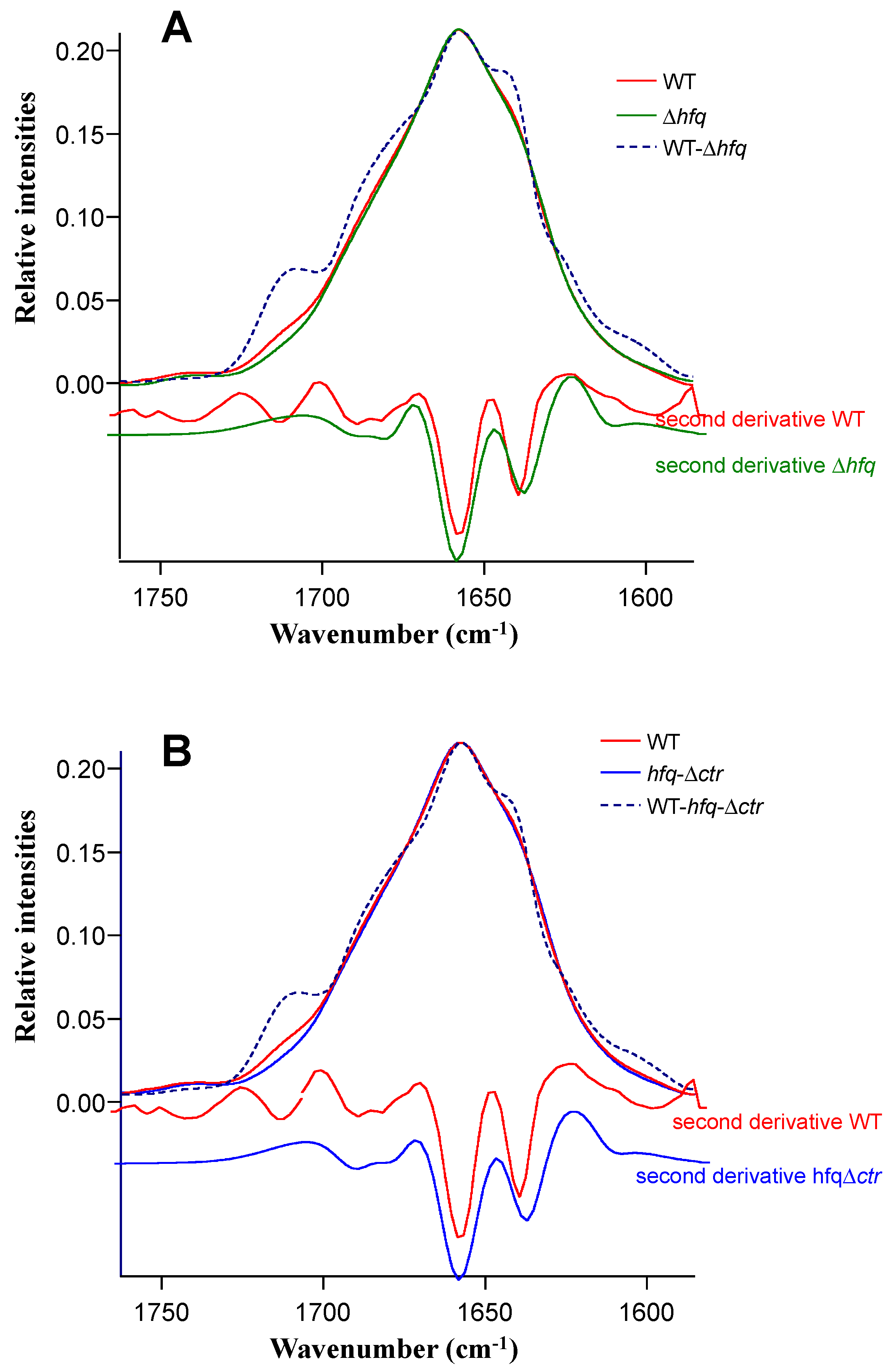
2.1. FTIR spectra and Variance Between Hfq Mutated Strains
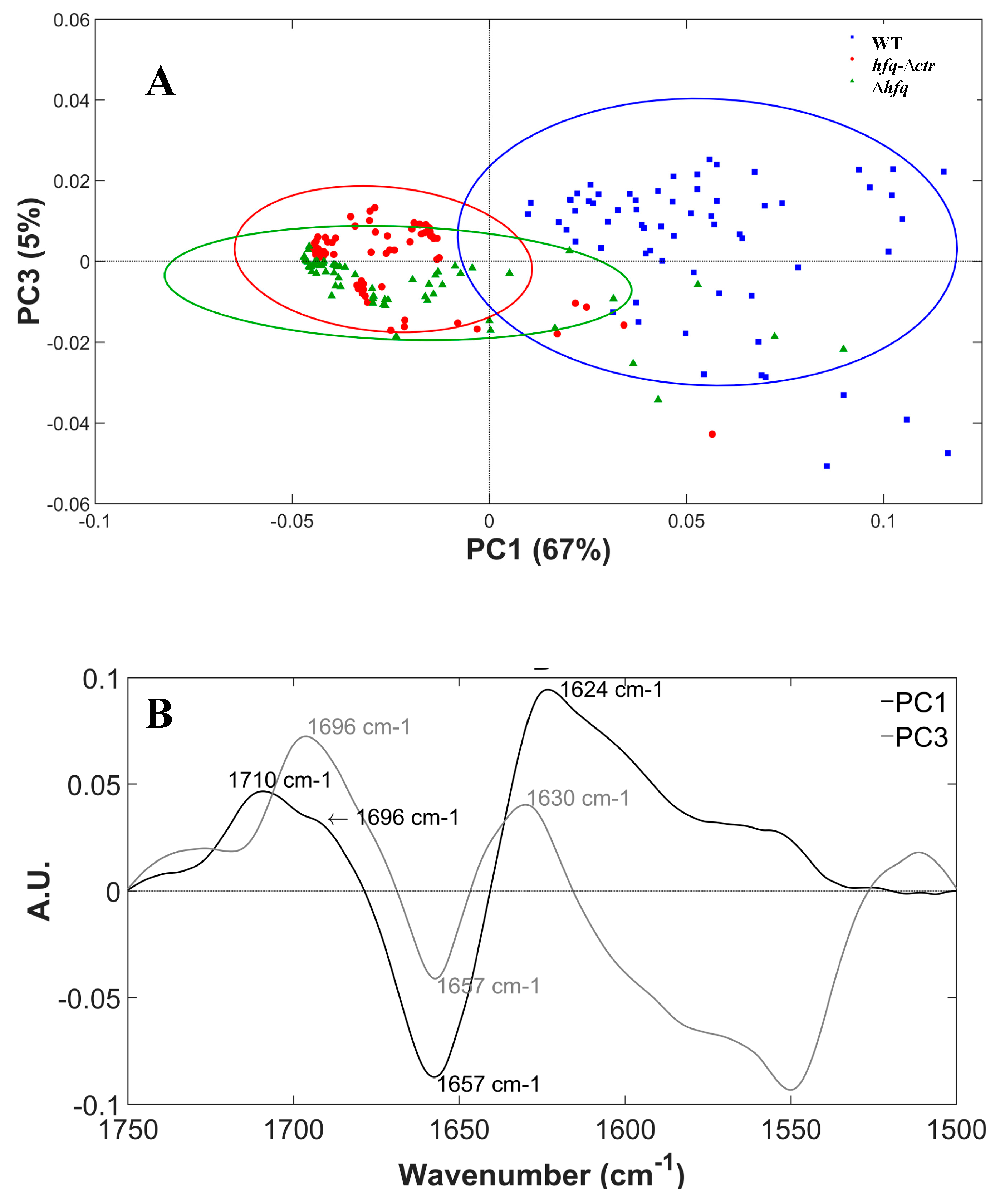
2.2. Multivariate Analysis Results
3. Materials and Methods
3.1. E. coli Strains
3.2. FTIRSpectroscopy and Principal Components Analysis
3.3. Synchrotron Radiation Circular Dichroism (SRCD)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Poole, K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. 2012, 20, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.F.; Han, K.; Chandler, C.E.; Tjaden, B.; Ernst, R.K.; Lory, S. Probing the sRNA regulatory landscape of P. aeruginosa: Post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility. Mol. Microbiol. 2017, 106, 919–937. [Google Scholar] [CrossRef] [PubMed]
- Mitarai, N.; Benjamin, J.A.; Krishna, S.; Semsey, S.; Csiszovszki, Z.; Masse, E.; Sneppen, K. Dynamic features of gene expression control by small regulatory RNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 10655–10659. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Sun, Y.; Miyakoshi, M.; Vanderpool, C.K.; Vogel, J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 2013, 153, 426–437. [Google Scholar] [CrossRef]
- Salvail, H.; Masse, E. Regulating iron storage and metabolism with RNA: An overview of posttranscriptional controls of intracellular iron homeostasis. Wiley Interdiscip. Rev. RNA 2012, 3, 26–36. [Google Scholar] [CrossRef]
- Sittka, A.; Pfeiffer, V.; Tedin, K.; Vogel, J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007, 63, 193–217. [Google Scholar] [CrossRef]
- Bardill, J.P.; Zhao, X.; Hammer, B.K. The Vibrio cholerae quorum sensing response is mediated by Hfq-dependent sRNA/mRNA base pairing interactions. Mol. Microbiol. 2011, 80, 1381–1394. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.; Vogel, J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010, 13, 24–33. [Google Scholar] [CrossRef]
- Kendall, M.M.; Gruber, C.C.; Rasko, D.A.; Hughes, D.T.; Sperandio, V. Hfq virulence regulation in enterohemorrhagic Escherichia coli O157:H7 strain 86-24. J. Bacteriol. 2011, 193, 6843–6851. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef]
- Majdalani, N.; Vanderpool, C.K.; Gottesman, S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 93–113. [Google Scholar] [CrossRef]
- Aiba, H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007, 10, 134–139. [Google Scholar] [CrossRef]
- Hwang, W.; Arluison, V.; Hohng, S. Dynamic competition of DsrA and rpoS fragments for the proximal binding site of Hfq as a means for efficient annealing. Nucleic Acids Res. 2011, 39, 5131–5139. [Google Scholar] [CrossRef]
- Arluison, V.; Hohng, S.; Roy, R.; Pellegrini, O.; Regnier, P.; Ha, T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007, 35, 999–1006. [Google Scholar] [CrossRef]
- Battesti, A.; Majdalani, N.; Gottesman, S. The RpoS-Mediated General Stress Response in Escherichia coli (*). Annu. Rev. Microbiol. 2011, 65, 189–213. [Google Scholar] [CrossRef]
- Cayrol, B.; Fortas, E.; Martret, C.; Cech, G.; Kloska, A.; Caulet, S.; Barbet, M.; Trepout, S.; Marco, S.; Taghbalout, A.; et al. Riboregulation of the bacterial actin-homolog MreB by DsrA small noncoding RNA. Integr. Biol. Quant. Biosci. Nano Macro 2015, 7, 128–141. [Google Scholar] [CrossRef]
- Mandin, P.; Gottesman, S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010, 29, 3094–3107. [Google Scholar] [CrossRef]
- Guillier, M.; Gottesman, S.; Storz, G. Modulating the outer membrane with small RNAs. Genes Dev. 2006, 20, 2338–2348. [Google Scholar] [CrossRef]
- Vogt, S.L.; Raivio, T.L. Hfq reduces envelope stress by controlling expression of envelope-localized proteins and protein complexes in enteropathogenic Escherichia coli. Mol. Microbiol. 2014, 92, 681–697. [Google Scholar] [CrossRef]
- Feliciano, J.R.; Grilo, A.M.; Guerreiro, S.I.; Sousa, S.A.; Leitao, J.H. Hfq: A multifaceted RNA chaperone involved in virulence. Future Microbiol. 2016, 11, 137–151. [Google Scholar] [CrossRef]
- Diestra, E.; Cayrol, B.; Arluison, V.; Risco, C. Cellular electron microscopy imaging reveals the localization of the Hfq protein close to the bacterial membrane. PLoS ONE 2009, 4, e8301. [Google Scholar] [CrossRef]
- Fortas, E.; Piccirilli, F.; Malabirade, A.; Militello, V.; Trepout, S.; Marco, S.; Taghbalout, A.; Arluison, V. New insight into the structure and function of Hfq C-terminus. Biosci. Rep. 2015, 35. [Google Scholar] [CrossRef]
- Taghbalout, A.; Yang, Q.; Arluison, V. The Escherichia coli RNA processing and degradation machinery is compartmentalized within an organized cellular network. Biochem. J. 2014, 458, 11–22. [Google Scholar] [CrossRef]
- Brennan, R.G.; Link, T.M. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007, 10, 125–133. [Google Scholar] [CrossRef]
- Wilusz, C.J.; Wilusz, J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013, 10, 592–601. [Google Scholar] [CrossRef]
- Partouche, D.; Malabirade, A.; Bizien, T.; Velez, M.; Trepout, S.; Marco, S.; Militello, V.; Sandt, C.; Wien, F.; Arluison, V. Techniques to Analyze sRNA Protein Cofactor Self-Assembly In Vitro. Methods Mol. Biol. 2018, 1737, 321–340. [Google Scholar] [CrossRef]
- Malabirade, A.; Partouche, D.; El Hamoui, O.; Turbant, F.; Geinguenaud, F.; Recouvreux, P.; Bizien, T.; Busi, F.; Wien, F.; Arluison, V. Revised role for Hfq bacterial regulator on DNA topology. Sci. Rep. 2018, 8, 16792. [Google Scholar] [CrossRef]
- Arluison, V.; Mura, C.; Guzman, M.R.; Liquier, J.; Pellegrini, O.; Gingery, M.; Regnier, P.; Marco, S. Three-dimensional Structures of Fibrillar Sm Proteins: Hfq and Other Sm-like Proteins. J. Mol. Biol. 2006, 356, 86–96. [Google Scholar] [CrossRef]
- Arluison, V.; Folichon, M.; Marco, S.; Derreumaux, P.; Pellegrini, O.; Seguin, J.; Hajnsdorf, E.; Regnier, P. The C-terminal domain of Escherichia coli Hfq increases the stability of the hexamer. Eur. J. Biochem. 2004, 271, 1258–1265. [Google Scholar] [CrossRef]
- Malabirade, A.; Morgado-Brajones, J.; Trepout, S.; Wien, F.; Marquez, I.; Seguin, J.; Marco, S.; Velez, M.; Arluison, V. Membrane association of the bacterial riboregulator Hfq and functional perspectives. Sci. Rep. 2017, 7, 10724. [Google Scholar] [CrossRef]
- Malabirade, A.; Jiang, K.; Kubiak, K.; Diaz-Mendoza, A.; Liu, F.; van Kan, J.A.; Berret, J.F.; Arluison, V.; van der Maarel, J.R.C. Compaction and condensation of DNA mediated by the C-terminal domain of Hfq. Nucleic Acids Res. 2017, 45, 7299–7308. [Google Scholar] [CrossRef]
- Beich-Frandsen, M.; Vecerek, B.; Konarev, P.V.; Sjoblom, B.; Kloiber, K.; Hammerle, H.; Rajkowitsch, L.; Miles, A.J.; Kontaxis, G.; Wallace, B.A.; et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011. [Google Scholar] [CrossRef]
- Navarra, G.; Leone, M.; Militello, V. Thermal aggregation of beta-lactoglobulin in presence of metal ions. Biophys. Chem. 2007, 131, 52–61. [Google Scholar] [CrossRef]
- Navarra, G.; Troia, F.; Militello, V.; Leone, M. Characterization of the nucleation process of lysozyme at physiological pH: Primary but not sole process. Biophys. Chem. 2013, 177–178, 24–33. [Google Scholar] [CrossRef]
- Tatulian, S.A. Structural characterization of membrane proteins and peptides by FTIR and ATR-FTIR spectroscopy. Methods Mol. Biol. 2013, 974, 177–218. [Google Scholar] [CrossRef]
- Zandomeneghi, G.; Krebs, M.R.; McCammon, M.G.; Fandrich, M. FTIR reveals structural differences between native beta-sheet proteins and amyloid fibrils. Protein Sci. 2004, 13, 3314–3321. [Google Scholar] [CrossRef]
- Aguilera, P.; Marcoleta, A.; Lobos-Ruiz, P.; Arranz, R.; Valpuesta, J.M.; Monasterio, O.; Lagos, R. Identification of Key Amino Acid Residues Modulating Intracellular and In vitro Microcin E492 Amyloid Formation. Front. Microbiol. 2016, 7, 35. [Google Scholar] [CrossRef]
- Gasset-Rosa, F.; Coquel, A.-S.; Moreno-Del Alamo, M.; Chen, P.; Song, X.; Serrano, A.M.; Fernandez-Tresguerres, M.E.; Moreno-Diaz de la Espina, S.; Lindner, A.B.; Giraldo, R. Direct assessment in bacteria of prionoid propagation and phenotype selection by Hsp70 chaperone. Mol. Microbiol. 2014, 91, 1070–1087. [Google Scholar] [CrossRef]
- Vetri, V.; Carrotta, R.; Picone, P.; Di Carlo, M.; Militello, V. Concanavalin A aggregation and toxicity on cell cultures. Biochim. Biophys. Acta 2010, 1804, 173–183. [Google Scholar] [CrossRef]
- Stains, C.I.; Mondal, K.; Ghosh, I. Molecules that target beta-amyloid. ChemMedChem 2007, 2, 1674–1692. [Google Scholar] [CrossRef]
- D’Amico, M.; Di Carlo, M.G.; Groenning, M.; Militello, V.; Vetri, V.; Leone, M. Thioflavin T Promotes Aβ(1−40) Amyloid Fibrils Formation. Biophys. Chem. 2012, 3, 1596–1601. [Google Scholar] [CrossRef]
- Zhou, Y.; Blanco, L.P.; Smith, D.R.; Chapman, M.R. Bacterial amyloids. Methods Mol. Biol. 2012, 849, 303–320. [Google Scholar] [CrossRef]
- Zambrano, N.; Guichard, P.P.; Bi, Y.; Cayrol, B.; Marco, S.; Arluison, V. Involvement of HFq protein in the post-transcriptional regulation of E. coli bacterial cytoskeleton and cell division proteins. Cell Cycle 2009, 8, 2470–2472. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Ragunathan, P.T.; Fei, J.; Vanderpool, C.K. A Prophage-Encoded Small RNA Controls Metabolism and Cell Division in Escherichia coli. mSystems 2016, 1, e00021-15. [Google Scholar] [CrossRef]
- Serra, D.O.; Mika, F.; Richter, A.M.; Hengge, R. The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and downregulating the biofilm regulator CsgD via the sigma(E)-dependent sRNA RybB. Mol. Microbiol. 2016, 101, 136–151. [Google Scholar] [CrossRef]
- Andreassen, P.R.; Pettersen, J.S.; Szczerba, M.; Valentin-Hansen, P.; Moller-Jensen, J.; Jorgensen, M.G. sRNA-dependent control of curli biosynthesis in Escherichia coli: McaS directs endonucleolytic cleavage of csgD mRNA. Nucleic Acids Res. 2018, 46, 6746–6760. [Google Scholar] [CrossRef]
- Terry, C.; Harniman, R.L.; Sells, J.; Wenborn, A.; Joiner, S.; Saibil, H.R.; Miles, M.J.; Collinge, J.; Wadsworth, J.D.F. Structural features distinguishing infectious ex vivo mammalian prions from non-infectious fibrillar assemblies generated in vitro. Sci. Rep. 2019, 9, 376. [Google Scholar] [CrossRef]
- Chirgadze, Y.N.; Nevskaya, N.A. Infrared spectra and resonance interaction of amide-I vibration of the paraellel-chain pleated sheets. Biopolymers 1976, 15, 627–636. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyaki, E.; Kun, J.; Moussong, E.; Lee, Y.H.; Goto, Y.; Refregiers, M.; Kardos, J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Ami, D.; Bonecchi, L.; Calì, S. FT-IR study of heterologous protein expression in recombinant Escherichia coli strains. Biochim. Biophys. Acta 2003, 624, 6–10. [Google Scholar] [CrossRef]
- Jiang, K.; Zhang, C.; Guttula, D.; Liu, F.; van Kan, J.A.; Lavelle, C.; Kubiak, K.; Malabirade, A.; Lapp, A.; Arluison, V.; et al. Effects of Hfq on the conformation and compaction of DNA. Nucleic Acids Res. 2015, 43, 4332–4341. [Google Scholar] [CrossRef]
- Refregiers, M.; Wien, F.; Ta, H.P.; Premvardhan, L.; Bac, S.; Jamme, F.; Rouam, V.; Lagarde, B.; Polack, F.; Giorgetta, J.L.; et al. DISCO synchrotron-radiation circular-dichroism endstation at SOLEIL. J. Synchrotron Radiat. 2012, 19, 831–835. [Google Scholar] [CrossRef]
- Partouche, D.; Turbant, F.; El Hamoui, O.; Campidelli, C.; Bombled, M.; Trepout, S.; Wien, F.; Arluison, V. Epigallocatechin Gallate Remodelling of Hfq Amyloid-Like Region Affects Escherichia coli Survival. Pathogens 2018, 7, 95. [Google Scholar] [CrossRef]
- Lees, J.G.; Smith, B.R.; Wien, F.; Miles, A.J.; Wallace, B.A. CDtool-an integrated software package for circular dichroism spectroscopic data processing, analysis, and archiving. Anal. Biochem. 2004, 332, 285–289. [Google Scholar] [CrossRef]
- Dazzi, A.; Prater, C.B. AFM-IR: Technology and Applications in Nanoscale Infrared Spectroscopy and Chemical Imaging. Chem. Rev. 2016. [Google Scholar] [CrossRef]

| Amide I Components (cm−1) | % | Assignment |
|---|---|---|
| Hfq full-length in vivo | ||
| 1614 | 2 | intermolecular β-sheet |
| 1625 | 5 | amyloid β-sheet |
| 1640 | 14.5 | β-sheet |
| 1656 | 31.5 | random coil |
| 1675 | 11 | turns/β-aggregated |
| 1687 | 13 | β-sheet |
| 1710 | 9 | side chains |
| Hfq-CTR in vivo | ||
| 1614 | 2 | intermolecular β-sheet |
| 1625 | 6.5 | amyloid β-sheet |
| 1640 | 12 | β-sheet |
| 1656 | 34 | random coil |
| 1674 | 13 | turns/β-aggregated |
| 1687 | 10 | β-sheet |
| 1710 | 10 | side chains |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Partouche, D.; Militello, V.; Gomez-Zavaglia, A.; Wien, F.; Sandt, C.; Arluison, V. In Situ Characterization of Hfq Bacterial Amyloid: A Fourier-Transform Infrared Spectroscopy Study. Pathogens 2019, 8, 36. https://doi.org/10.3390/pathogens8010036
Partouche D, Militello V, Gomez-Zavaglia A, Wien F, Sandt C, Arluison V. In Situ Characterization of Hfq Bacterial Amyloid: A Fourier-Transform Infrared Spectroscopy Study. Pathogens. 2019; 8(1):36. https://doi.org/10.3390/pathogens8010036
Chicago/Turabian StylePartouche, David, Valeria Militello, Andrea Gomez-Zavaglia, Frank Wien, Christophe Sandt, and Véronique Arluison. 2019. "In Situ Characterization of Hfq Bacterial Amyloid: A Fourier-Transform Infrared Spectroscopy Study" Pathogens 8, no. 1: 36. https://doi.org/10.3390/pathogens8010036
APA StylePartouche, D., Militello, V., Gomez-Zavaglia, A., Wien, F., Sandt, C., & Arluison, V. (2019). In Situ Characterization of Hfq Bacterial Amyloid: A Fourier-Transform Infrared Spectroscopy Study. Pathogens, 8(1), 36. https://doi.org/10.3390/pathogens8010036






