The Mosquito Immune System and the Life of Dengue Virus: What We Know and Do Not Know
Abstract
:1. Introduction
2. Innate Immunity
- Antiviral signaling pathways: Toll-like pathway, immune deficiency (IMD), and Janus kinase-signal transducer and activator of transcription (JAK-STAT) pathway.
- Complement-like proteins: These include Thioester-containing proteins (TEP); A. aegypti macroglobulin complement related factor (AaMCR) and A. aegypti homolog of scavenger receptor-C (AaSR-C).
- Small RNAs: These include small interfering RNA (siRNA), micro RNA (miRNA) (derived from the virus), and PIWI-interacting RNA (piRNA).
3. Antiviral Signaling Pathways
4. Organ-Specific Antiviral Strategies
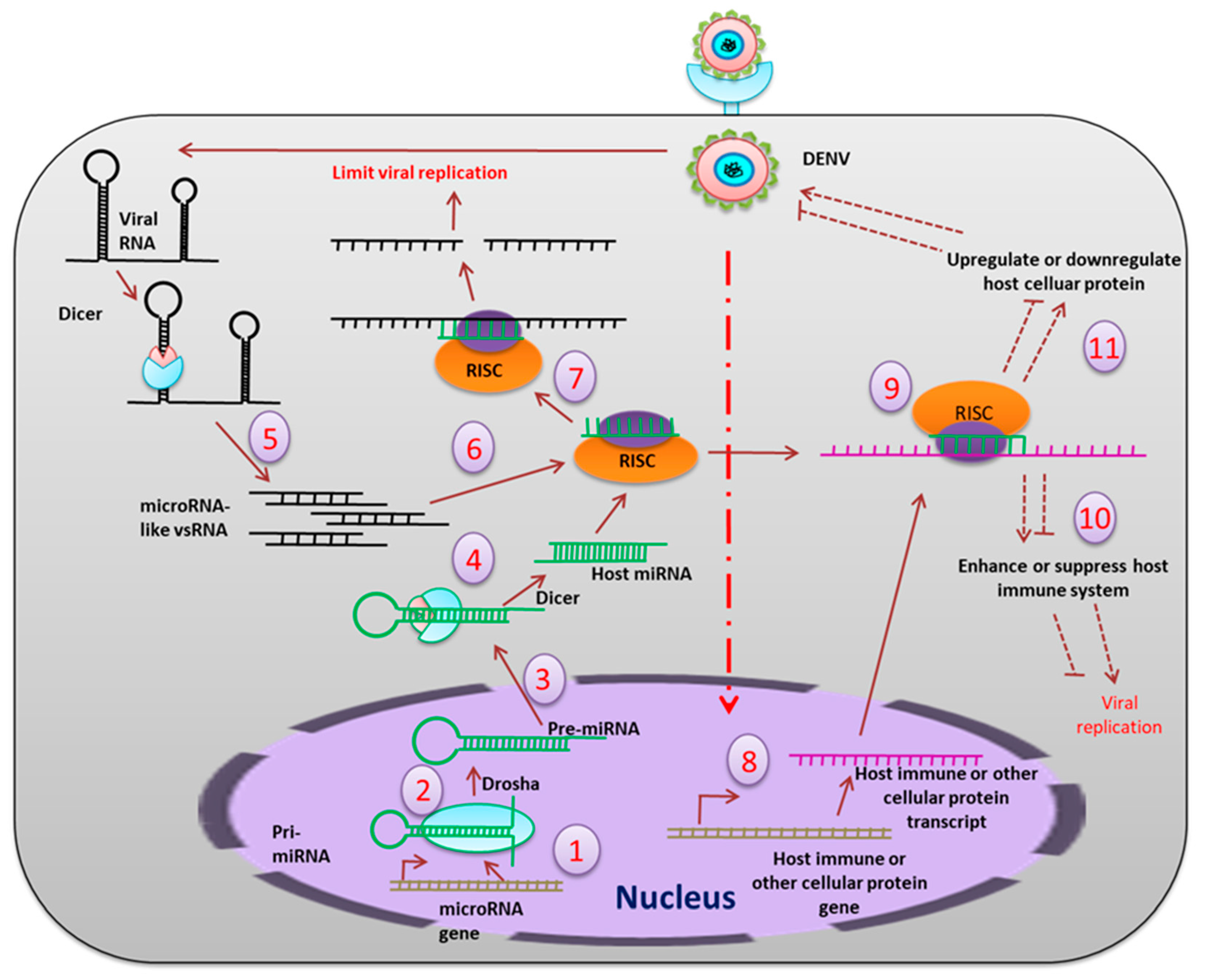
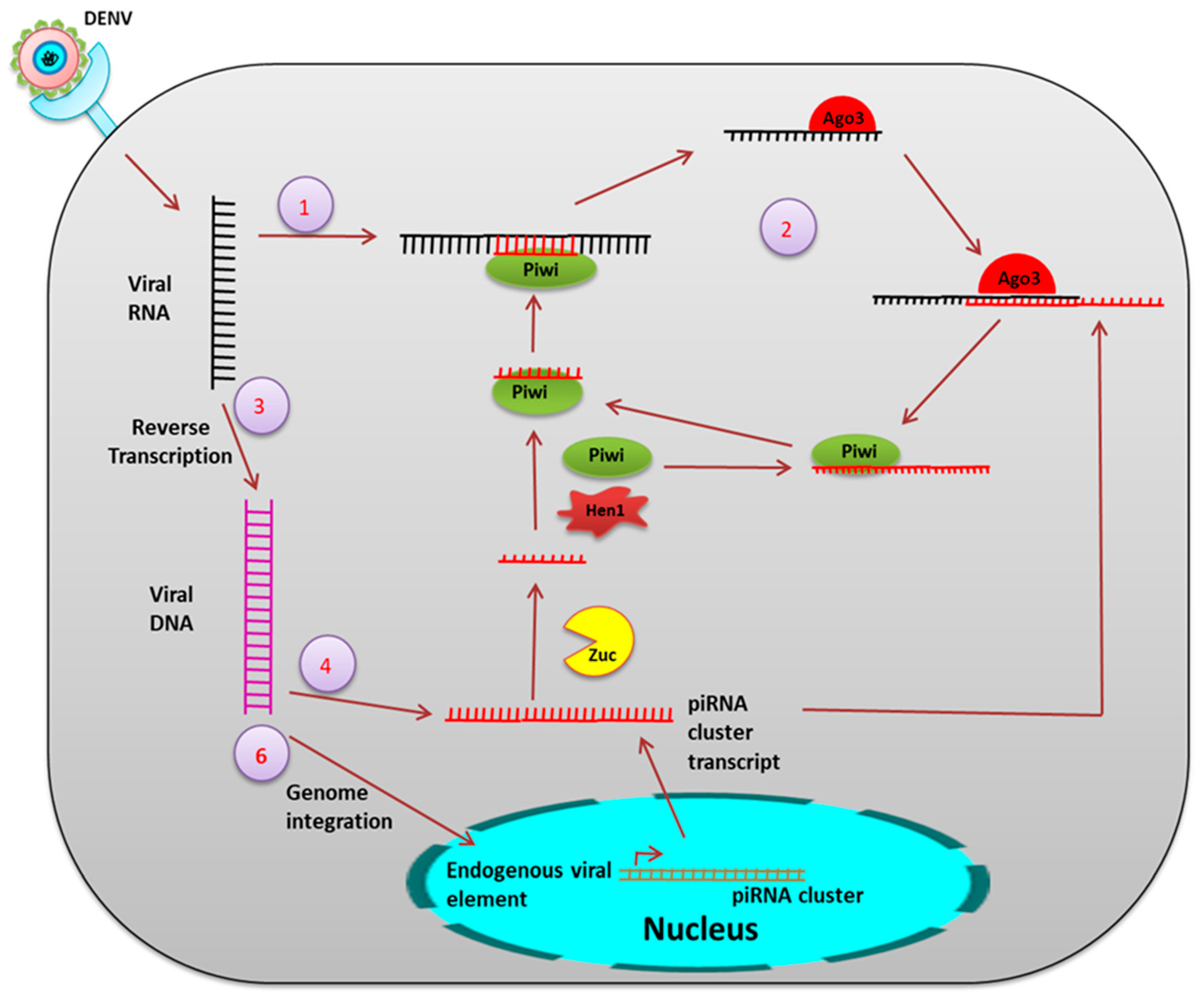
5. Small RNA Mediated Immunity
6. Microbial Community
7. Lipid Droplet (LD) and Immunity
8. Adaptive Immunity
9. What is Next?
Author Contributions
Conflicts of Interest
References
- Frolova, E.I.; Fayzulin, R.Z.; Cook, S.H.; Griffin, D.E.; Rice, C.M.; Frolov, I. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis Virus Infection. J. Virol. 2002, 76, 11254–11264. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Dimopoulos, G. Dengue virus inhibits immune responses in Aedes aegypti Cells. PLoS ONE 2010, 5, e10678. [Google Scholar] [CrossRef]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti Toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Dimopoulos, G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Dev. Comp. Immunol. 2010, 34, 625–629. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.N.; Tauszig-Delamasure, S.; Hoffmann, J.A.; Lelièvre, E.; Gascan, H.; Ray, K.P.; Morse, M.A.; Imler, J.; Gay, N.J. Binding of the Drosophila cytokine Spätzle to Toll is direct and establishes signaling. Nat. Immunol. 2003, 4, 794–800. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Muturi, E.J.; Barletta, A.B.; Rooney, A.P. The Aedes aegypti IMD pathway is a critical component of the mosquito antifungal immune response. Dev. Comp. Immunol. 2019, 95, 1–9. [Google Scholar] [CrossRef]
- Shin, S.W.; Kokoza, V.; Ahmed, A.; Raikhel, A.S. Characterization of three alternatively spliced isoforms of the Rel/NF- B transcription factor Relish from the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2002, 99, 9978–9983. [Google Scholar] [CrossRef]
- Georgel, P.; Naitza, S.; Kappler, C.; Ferrandon, D.; Zachary, D.; Swimmer, C.; Kopczynski, C.; Duyk, G.; Reichhart, J.M.; Hoffmann, J.A. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 2001, 1, 503–514. [Google Scholar] [CrossRef]
- Paradkar, P.N.; Trinidad, L.; Voysey, R.; Duchemin, J.B.; Walker, P.J. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 18915–18920. [Google Scholar] [CrossRef] [PubMed]
- Deddouche, S.; Matt, N.; Budd, A.; Mueller, S.; Kemp, C.; Galiana-Arnoux, D.; Dostert, C.; Antoniewski, C.; Hoffmann, J.A.; Imler, J.L. The DExD/H-boxhelicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat. Immunol 2008, 9, 1425–1432. [Google Scholar] [CrossRef]
- Luplertlop, N.; Surasombatpattana, P.; Patramool, S.; Dumas, E.; Wasinpiyamongkol, L.; Saune, L.; Hamel, R.; Bernard, E.; Sereno, D.; Thomas, F.; et al. Induction of a peptide with activity against a broad spectrum of pathogens in the Aedes aegypti salivary gland, following infection with dengue virus. PLoS Pathog. 2011, 7, e1001252. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.; Jupatanakul, N.; Ramirez, J.L.; Kang, S.; Romero-Vivas, C.M.; Mohammed, H.; Dimopoulos, G. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS Negl. Trop. Dis. 2013, 7, e2295. [Google Scholar] [CrossRef] [PubMed]
- Silverman, N.; Zhou, R.; Erlich, R.L.; Hunter, M.; Bernstein, E.; Schneider, D.; Maniatis, T. Immune activation of NF-κB and JNK requires Drosophila TAK1. J. Biol. Chem. 2003, 278, 48928–48934. [Google Scholar] [CrossRef] [PubMed]
- Choy, M.M.; Sessions, O.M.; Gubler, D.J.; Ooi, E.E. Production of infectious dengue virus in Aedes aegypti is dependent on the ubiquitin proteasome pathway. PLoS Negl. Trop. Dis. 2015, 9, e0004227. [Google Scholar] [CrossRef] [PubMed]
- Troupin, A.; Londono-Renteria, B.; Conway, M.J.; Cloherty, E.; Jameson, S.; Higgs, S.; Vanlandingham, D.L.; Fikrig, E.; Colpitts, T.M. A novel mosquito ubiquitin targets viral envelope protein for degradation and reduces virion production during dengue virus infection. Biochim. Biophys. Acta 2016, 1860, 1898–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souza-Neto, J.A.; Sim, S.; Dimopoulos, G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. USA 2009, 106, 17841–17846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, N.; Mueller, C.R.; Fuchs, J.F.; McElroy, K.; Wessely, V.; Higgs, S.; Christensen, B.M. Evaluation of the function of a type I peritrophic matrix as a physical barrier for midgut epithelium invasion by mosquito-borne pathogens in Aedes aegypti. Vector-Borne Zoonotic Dis. 2008, 8, 701–712. [Google Scholar] [CrossRef]
- Houk, E.; Obie, F.; Hardy, J. Peritrophic membrane formation and the midgut barrier to arboviral infection in the mosquito, Culex tarsalis Coquillett (Insecta, Diptera). Acta Trop. 1979, 36, 39–45. [Google Scholar]
- Beaty, B.J.; Bishop, D.H. Bunyavirus-vector interactions. Virus Res. 1988, 10, 289–301. [Google Scholar] [CrossRef]
- Black, I.V.W.C.; Bennett, K.E.; Gorrochótegui-Escalante, N.; Barillas-Mury, C.V.; Fernández-Salas, I.; de Lourdes Muñoz, M.; Farfán-Alé, J.A.; Olson, K.E.; Beaty, B.J. Flavivirus Susceptibility in Aedes aegypti. Arch. Med. Res. 2002, 33, 379–388. [Google Scholar] [CrossRef]
- Sánchez-Vargas, I.; Scott, J.C.; Poole-Smith, B.K.; Franz, A.W.E.; Barbosa-Solomieu, V.; Wilusz, J.; Olson, K.E.; Blair, C.D. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito’s RNA interference pathway. PLoS Pathog. 2009, 5, e1000299. [Google Scholar] [CrossRef]
- Ramos-Castañeda, J.; González, C.; Jiménez, M.A.; Duran, J.; Hernández-Martínez, S.; Rodríguez, M.H.; Lanz-Mendoza, H. Effect of nitric oxide on dengue virus replication in Aedes aegypti and Anopheles albimanus. Intervirology 2008, 51, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Liu, L.; Wang, P.; Zhang, Y.; Zhao, Y.O.; Colpitts, T.M.; Feitosa, F.; Anderson, J.F.; Fikrig, E. An in vivo transfection approach elucidates a role for Aedes aegypti thioester-containing proteins in flaviviral infection. PLoS ONE 2011, 6, e22786. [Google Scholar] [CrossRef]
- Xiao, X.; Liu, Y.; Zhang, X.; Wang, J.; Li, Z.; Pang, X.; Wang, P.; Cheng, G. Complement-related proteins control the flavivirus infection of Aedes aegypti by inducing antimicrobial peptides. PLoS Pathog. 2014, 10, e1004027. [Google Scholar] [CrossRef]
- Kramer, L.D.; Hardy, J.L.; Presser, S.B.; Houk, E.J. Dissemination barriers for western equine encephalomyelitis virus in Culex tarsalis infected after ingestion of low viral doses. Am. J. Trop. Med. Hyg. 1981, 30, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Romoser, W.S.; Turell, M.J.; Lerdthusnee, K.; Neira, M.; Dohm, D.; Luldwig, G.; Wasieloski, L. Pathogenesis of Rift Valley fever virus in mosquitoes-tracheal conduits and the basal lamina as an extra-cellular barrier. Arch. Virol. Suppl. 2005, 19, 89–100. [Google Scholar]
- Grimstad, P.R.; Paulson, S.L.; Craig, G.B., Jr. Vector competence of Aedes hendersoni (Diptera: Culicidae) for La Crosse virus and evidence of a salivary-gland escape barrier. J. Med. Entomol. 1985, 22, 447–453. [Google Scholar] [CrossRef]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, M.; Matsuzaki, F.; Nabeshima, Y.-I.; Hama, C. hikaru genki, a CNS-specific gene identified by abnormal locomotion in Drosophila, encodes a novel type of protein. Neuron 1993, 10, 395–407. [Google Scholar] [CrossRef]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-related genes and gene families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef]
- Matsuyama, K.; Natori, S. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J. Biol. Chem. 1988, 263, 17112–17116. [Google Scholar] [PubMed]
- Wimley, W.C.; Selsted, M.E.; White, S.H. Interactions between human defensins and lipid bilayers: Evidence for formation of multimeric pores. Protein Sci. 1994, 3, 1362–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A.J.; Beazley, W.D.; Bibby, M.C.; Devine, D.A. Antimicrobial activity of cecropins. J. Antimicrobiol. Chemother. 1996, 37, 1077–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Guo, C.; Huang, Y.; Zhang, X.; Chen, Y. Inhibition of porcine reproductive and respiratory syndrome virus by Cecropin D in vitro. Infect. Genet. Evol. 2015, 34, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Huang, Y.; Cong, P.; Liu, X.; Chen, Y.; He, Z. Cecropin P1 inhibits porcine reproductive and respiratory syndrome virus by blocking attachment. BMC Microbiol. 2014, 14, 273. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Kingsolver, M.B.; Avadhanula, V.; Hardy, R.W. An antiviral role for antimicrobial peptides during the arthropod response to Alphavirus replication. J. Virol. 2013, 87, 4272–4280. [Google Scholar] [CrossRef] [PubMed]
- Gaines, P.J.; Olson, K.E.; Higgs, S.; Powers, A.M.; Beaty, B.J.; Blair, C.D. Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome. J. Virol. 1996, 70, 2132–2137. [Google Scholar] [PubMed]
- Caplen, N.J.; Zheng, Z.; Falgout, B.; Morgan, R.A. Inhibition of viral gene expression and replication in mosquito cells by dsRNA-triggered RNA interference. Mol. Ther. 2002, 6, 243–251. [Google Scholar] [CrossRef]
- Adelman, Z.N.; Blair, C.D.; Carlson, J.O.; Beaty, B.J.; Olson, K.E. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol. Biol. 2001, 10, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Hess, A.M.; Prasad, A.N.; Ptitsyn, A.; Ebel, G.D.; Olson, K.E.; Barbacioru, C.; Monighetti, C.; Campbell, C.L. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 2011, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hong, H.; Yue, J.; Wu, Y.; Li, X.; Jiang, L.; Li, L.; Li, Q.; Gao, G.; Yang, X. Inhibitory effect of small interfering RNA on dengue virus replication in mosquito cells. Virol. J. 2010, 7, 270. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.-X.; Ding, S.W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Walker, T.; Oneill, S.L.; Asgari, S. Blood meal induced microRNA regulates development and immune associated genes in the Dengue mosquito vector, Aedes aegypti. Insect Biochem. Mol. Biol. 2013, 43, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Wu, J.; Zheng, P.; Li, Y.; Zheng, X.; Puthiyakunnon, S.; Tu, Z.; Chen, X.G. The expression profile of Aedes albopictus miRNAs is altered by dengue virus serotype-2 infection. Cell Biosci. 2015, 5, 16. [Google Scholar] [CrossRef]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; Nova-Ocampo, M.A.D.; Ángel, R.M.D.; Salas-Benito, J.S. Analysis of the miRNA profile in C6/36 cells persistently infected with dengue virus type 2. Virus Res. 2017, 232, 139–151. [Google Scholar] [CrossRef]
- Hussain, M.; Asgari, S. MicroRNA-like viral small RNA from Dengue virus 2 autoregulates its replication in mosquito cells. Proc. Natl. Acad. Sci. USA 2014, 111, 2746–2751. [Google Scholar] [CrossRef] [Green Version]
- Yen, P.-S.; James, A.; Li, J.-C.; Chen, C.-H.; Failloux, A.-B. Synthetic miRNAs induce dual arboviral-resistance phenotypes in the vector mosquito Aedes aegypti. Commun. Biol. 2018, 1, 11. [Google Scholar] [CrossRef]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Ipsaro, J.J.; Haase, A.D.; Knott, S.R.; Joshua-Tor, L.; Hannon, G.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012, 491, 279–283. [Google Scholar] [CrossRef] [Green Version]
- Kawaoka, S.; Izumi, N.; Katsuma, S.; Tomari, Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell 2011, 43, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of PIWI- interacting RNAs at their 3′ ends. Genes Dev. 2007, 21, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009, 137, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Homolka, D.; Pandey, R.R.; Goriaux, C.; Brasset, E.; Vaury, C.; Sachidanandam, R.; Fauvarque, M.O.; Pillai, R.S. PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep. 2015, 12, 418–428. [Google Scholar] [CrossRef]
- Scott, J.C.; Brackney, D.E.; Campbell, C.L.; Bondu-Hawkins, V.; Hjelle, B.; Ebel, G.D.; Oslon, K.E.; Blair, C.D. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and incompetent mosquito cells. PLoS Negl. Trop. Dis. 2010, 4, e848. [Google Scholar] [CrossRef] [PubMed]
- Miesen, P.; Ivens, A.; Buck, A.H.; Rij, R.P.V. Small RNA profiling in dengue virus 2-infected Aedes mosquito cells reveals viral piRNAs and novel host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, B.; Liu, P.; Li, J.; Chen, X.; Gu, J. piRNA profiling of dengue virus type 2-infected Asian tiger mosquito and midgut tissues. Viruses 2018, 10, 213. [Google Scholar] [CrossRef]
- Palatini, U.; Miesen, P.; Carballar-Lejarazu, R.; Ometto, L.; Rizzo, E.; Tu, Z.; van Rij, R.P.; Bonizzoni, M. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genom. 2017, 18, 512. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Souza-Neto, J.; Torres Cosme, R.; Rovira, J.; Ortiz, A.; Pascale, J.M.; Dimopoulos, G. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 2012, 6, e1561. [Google Scholar] [CrossRef]
- Ramirez, J.L.; Short, S.M.; Bahia, A.C.; Saraiva, R.G.; Dong, Y.; Kang, S.; Tripathi, A.; Mlambo, G.; Dimopoulos, G. Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PLoS Pathog. 2014, 10, e1004398. [Google Scholar] [CrossRef]
- Angleró-Rodríguez, Y.I.; Talyuli, O.A.; Blumberg, B.J.; Kang, S.; Demby, C.; Shields, A.; Carlson, J.; Jupatanakul, N.; Dimopoulos, G. An Aedes aegypti-associated fungus increases susceptibility to dengue virus by modulating gut trypsin activity. Elife 2017, 6, e28844. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Xu, Y.; Lu, P.; Xie, Y.; Xi, Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 2010, 6, e1000833. [Google Scholar] [CrossRef] [PubMed]
- Mousson, L.; Zouache, K.; Arias-Goeta, C.; Raquin, V.; Mavingui, P.; Failloux, A.-B. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl. Trop. Dis. 2012, 6, e1989. [Google Scholar] [CrossRef]
- Ferguson, N.M.; Kien, D.T.H.; Clapham, H.; Aguas, R.; Trung, V.T.; Chau, T.N.; Popovici, J.; Ryan, P.A.; O’Neill, S.L.; McGraw, E.A.; et al. Modeling the impact on virus transmission of Wolbachia-mediated blocking of dengue virus infection of Aedes aegypti. Sci. Transl. Med. 2015, 7, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.; Johnson, P.H.; Moreira, L.A.; Iturbe-Ormaetxe, I.; Frentiu, F.D.; McMeniman, C.J.; Leong, Y.S.; Dong, Y.; Axford, J.; Kriesner, P.; et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nat. Lett. 2011, 476, 450–453. [Google Scholar] [CrossRef]
- Thomas, S.; Verma, J.; Woolfit, M.; O’Neill, S.L. Wolbachia-mediated virus blocking in mosquito cells is dependent on XRN1-mediated viral RNA degradation and influenced by viral replication rate. PLoS Pathog. 2018, 14, e1006879. [Google Scholar] [CrossRef]
- Barletta, A.B.; Alves, L.R.; Silva, M.C.; Sim, S.; Dimopoulos, G.; Liechocki, S.; Maya-Monteiro, C.M.; Sorgine, M.H. Emerging role of lipid droplets in Aedes aegypti immune response against bacteria and Dengue virus. Sci. Rep. 2016, 6, 19928. [Google Scholar] [CrossRef]
- Wu, M.; Sun, L.V.; Vamathevan, J.; Riegler, M.; Deboy, R.; Brownlie, J.C.; McGraw, E.A.; Martin, W.; Esser, C.; Ahmadinejad, N.; et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004, 2, E69. [Google Scholar] [CrossRef]
- Molloy, J.C.; Sommer, U.; Viant, M.R.; Sinkins, S.P. Wolbachia modulates lipid metabolism in Aedes albopictus mosquito cells. Appl. Environ. Microbiol. 2016, 82, 3109–3120. [Google Scholar] [CrossRef]
- Pan, X.; Zhou, G.; Wu, J.; Bian, G.; Lu, P.; Raikhel, A.S.; Xi, Z. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. USA 2011, 109, E23–E31. [Google Scholar] [CrossRef]
- Asad, S.; Parry, R.; Asgari, S. Upregulation of Aedes aegypti Vago1 by Wolbachia and its effect on dengue virus replication. Insect Biochem. Mol. Biol. 2018, 92, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Hussaina, M.; Frentiua, F.D.; Moreiraa, L.A.; O’Neilla, S.L.; Asgari, S. Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc. Natl. Acad. Sci. USA 2011, 108, 9250–9255. [Google Scholar]
- Zhang, R.; Zhu, Y.; Pang, X.; Xiao, X.; Zhang, R.; Cheng, G. Regulation of antimicrobial peptides in Aedes aegypti Aag2 cells. Front. Cell. Infect. Microbiol. 2017, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Taylor, H.E.; Dimopoulos, G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006, 4, e229. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.-C.; Tassetto, M.; Rij, R.P.V.; Goic, B.; Gausson, V.; Berry, B.; Jacquier, C.; Antoniewski, C.; Andino, R. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 2009, 458, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, Z.J.; Dolan, P.T.; Kunitomi, M.; Tassetto, M.; Seetin, M.G.; Oh, S.; Heiner, C.; Paxinos, E.; Andino, R. The diversity, structure, and function of heritable adaptive immunity sequences in the Aedes aegypti genome. Curr. Biol. 2017, 27, 3511–3519. [Google Scholar] [CrossRef] [PubMed]
- Jupatanakul, N.; Sim, S.; Angleró-Rodríguez, Y.I.; Souza-Neto, J.; Das, S.; Poti, K.E.; Rossi, S.L.; Bergren, N.; Vasilakis, N.; Dimopuolos, G. Engineered Aedes aegypti JAK/STAT pathway-mediated immunity to dengue virus. PLoS Negl. Trop. Dis. 2017, 11, e0005187. [Google Scholar] [CrossRef] [PubMed]
- Fang, J. Ecology: A world without mosquitoes. Nature 2010, 466, 432–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukherjee, D.; Das, S.; Begum, F.; Mal, S.; Ray, U. The Mosquito Immune System and the Life of Dengue Virus: What We Know and Do Not Know. Pathogens 2019, 8, 77. https://doi.org/10.3390/pathogens8020077
Mukherjee D, Das S, Begum F, Mal S, Ray U. The Mosquito Immune System and the Life of Dengue Virus: What We Know and Do Not Know. Pathogens. 2019; 8(2):77. https://doi.org/10.3390/pathogens8020077
Chicago/Turabian StyleMukherjee, Debica, Sandeepan Das, Feroza Begum, Sweety Mal, and Upasana Ray. 2019. "The Mosquito Immune System and the Life of Dengue Virus: What We Know and Do Not Know" Pathogens 8, no. 2: 77. https://doi.org/10.3390/pathogens8020077
APA StyleMukherjee, D., Das, S., Begum, F., Mal, S., & Ray, U. (2019). The Mosquito Immune System and the Life of Dengue Virus: What We Know and Do Not Know. Pathogens, 8(2), 77. https://doi.org/10.3390/pathogens8020077




